A year ago in Nature Biotechnology, Becht et al. argued that UMAP preserved global structure better than t-SNE. Now @GCLinderman and me wrote a comment saying that their results were entirely due to the different initialization choices: biorxiv.org/content/10.110…. Thread. (1/n)
Here is the original paper: nature.com/articles/nbt.4… by @EtienneBecht @leland_mcinnes @EvNewell1 et al. They used three data sets and two quantitative evaluation metrics: (1) preservation of pairwise distances and (2) reproducibility across repeated runs. UMAP won 6/6. (2/10)
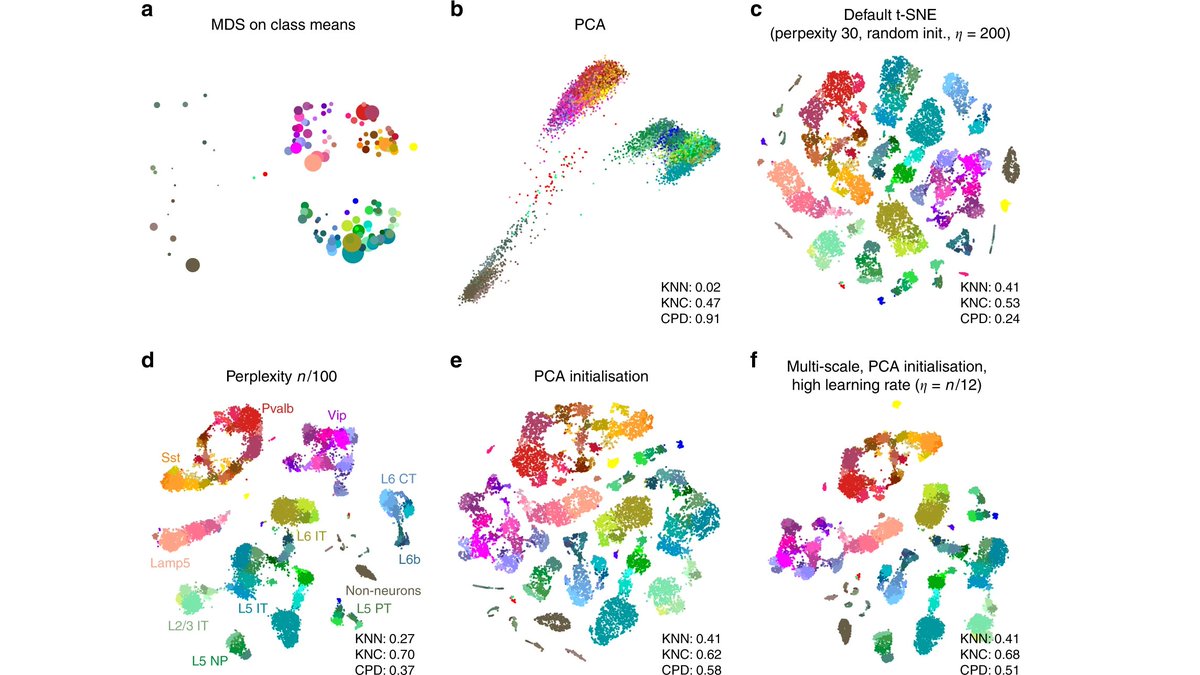
UMAP and t-SNE optimize different loss functions, but the implementations used in Becht et al. also used different default initialization choices: t-SNE was initialized randomly, whereas UMAP was initialized using the Laplacian eigenmaps (LE) embedding of the kNN graph. (3/10)
Were the results due to the different loss functions or due to the different initializations? George extended the code of Becht et al. to add UMAP with random initialization and t-SNE (using FIt-SNE) with PCA initialization to the benchmark comparison. This is the result. (4/10) 

Turns out, it was *entirely* due to initialization! UMAP with random initialization preserved global structure as poorly as t-SNE with random initialization, whereas t-SNE with informative (PCA) initialization performed as well as UMAP with informative (LE) initialization. (5/10)
This is particularly obvious for the reproducibility metric: of course if one runs t-SNE with random initialization and different random seeds, one can get very different global arrangements of clusters. People tend to think it is not true for UMAP, but we show that it is. (6/10)
In our view, the results of Becht et al. do not actually support the claim that UMAP preserves global structure better than t-SNE, which is how it's been cited in the field. The real lesson is that one should not be using random initialization for either of these methods. (7/10)
This is in agreement with the recommendation to use PCA initialization (rather than random initialization) for t-SNE made in the recent paper by @CellTypist and me:
https://twitter.com/hippopedoid/status/1206535867831083008. (8/10)
Just to be clear: this is *not* an attack on UMAP! I think UMAP is great :-) But I also think t-SNE is great. And there is plenty of room for further improvements and for better conceptual understanding of this whole family of embedding methods. (9/10)
But to decide which algorithm is more faithful to the single-cell data, further research is needed. Our Comment argues that Becht et al. paper does not answer that. (10/10)
A follow-up thread can be found here:
https://twitter.com/hippopedoid/status/1227731600647081984
UPDATE: Our comment was published in @NatureBiotech. There was no formal response from the authors so I assume we are all in agreement :-)
Free to read link: rdcu.be/cezFs
Journal link:
nature.com/articles/s4158…
Free to read link: rdcu.be/cezFs
Journal link:
nature.com/articles/s4158…
• • •
Missing some Tweet in this thread? You can try to
force a refresh