Our updated analysis on #SARSCoV2 clades in India, now peer-reviewed and published in OFID by @OxUniPress. In collaboration with @vinodscaria @IGIBSocial. Congratulations to the entire team.
doi.org/10.1093/ofid/o…
#Genomics #IndiaFightsCOVID19
doi.org/10.1093/ofid/o…
#Genomics #IndiaFightsCOVID19

I’ve never done one of these before, but I’m gonna make a thread of tweets explaining the key findings, and more importantly what our paper doesn’t show. 1/n
First and foremost, there is no conclusive evidence at present that any clade of SARS-CoV-2 is associated with increased mortality or severity of the disease. 2/n
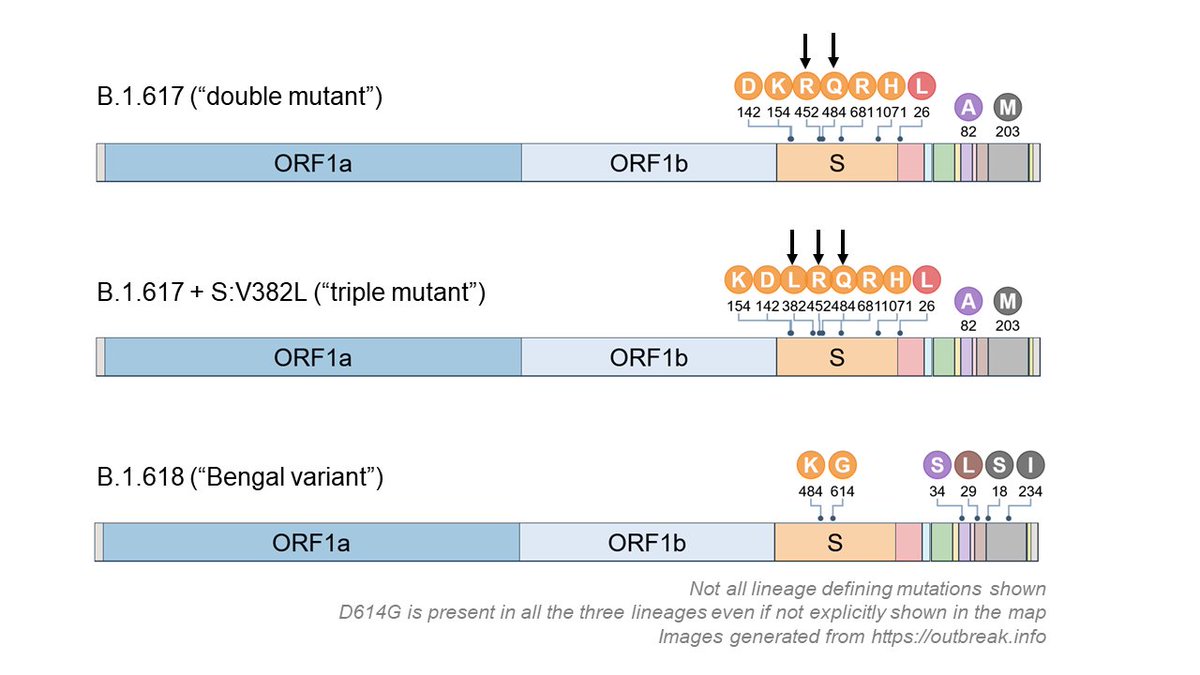
Our earlier bioRxiv study reported an uncategorized cluster of SARS-CoV-2 genomes which was responsible for the initial outbreak in India (Late Feb to April). We called it the I/A3i clade. 3/n
Viruses of I/A3i clade are identified by specific variations at 4 different places in their genomes. One of them is in RDRP - the viral enzyme needed for making more copies of viral RNA. 4/n 

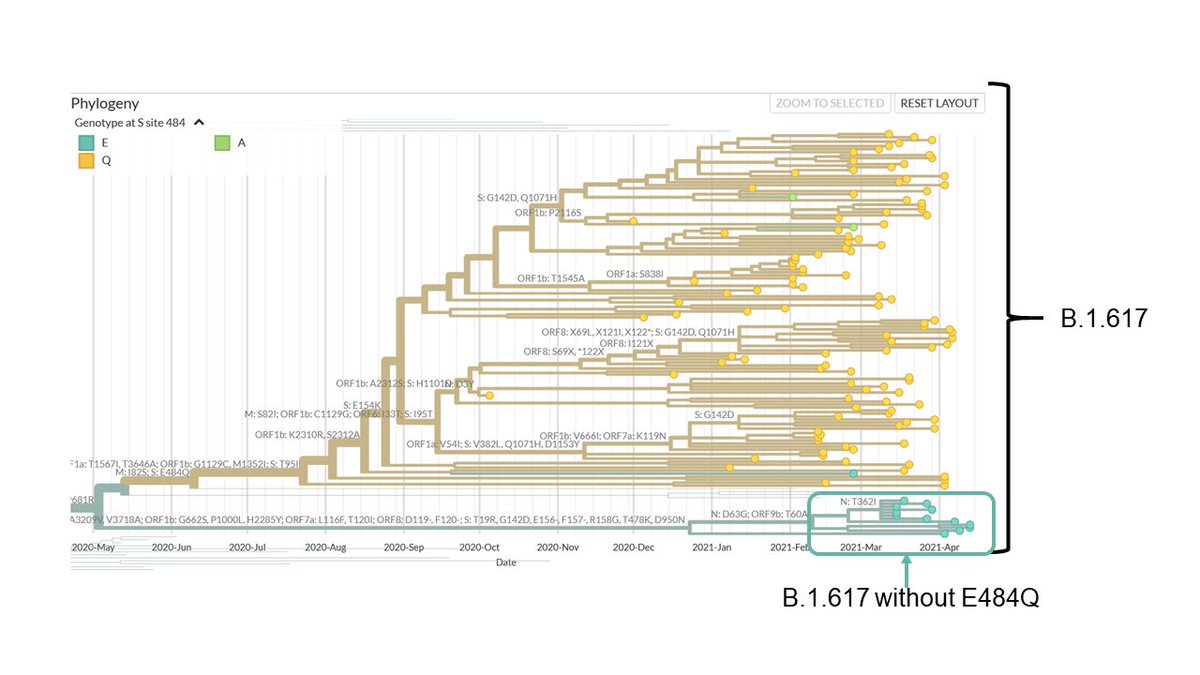
The RDRP variant is predicted to be bad for virus. If it is so, we expect A3i clade to slowly disappear, as other clades prevail. The updated analysis shows this - proportion of A3i clade reduced to ~18% now, compared to 41% when bioRxiv paper was submitted (early June). 5/n
The only place where we continue to see more of I/A3i clade is in Delhi. However, more data collected from recent time points would be required to see if the trend continues or Delhi also follows other states. 6/n
So, if not A3i, what is present in India? Almost 70% of isolates belong to the A2a clade, the globally prevalent clade. A2a, also known as the G clade or 20A/B/C, is the clade which carries the (in)famous D614G mutation in the Spike protein. 7/n
D614G mutation is shown to be more infectious in nature. Infectivity, not severity or death. Hence, it makes sense that viruses carrying this mutation will ultimately be over-represented everywhere, including India. This is good news because.. 8/n
If a vaccine/drug targeting this mutation is developed anywhere in the world, it will work with the same efficiency even here. 9/n
Finally, just to drive the point home, no clade at present is shown to be associated with increased severity or morbidity of COVID19. 10/10
• • •
Missing some Tweet in this thread? You can try to
force a refresh