There has been discussion over the past week about what the new @Apple M1 chip means for bioinformatics. Some have predicted the end of compbio on @Apple. Others are more optimistic.
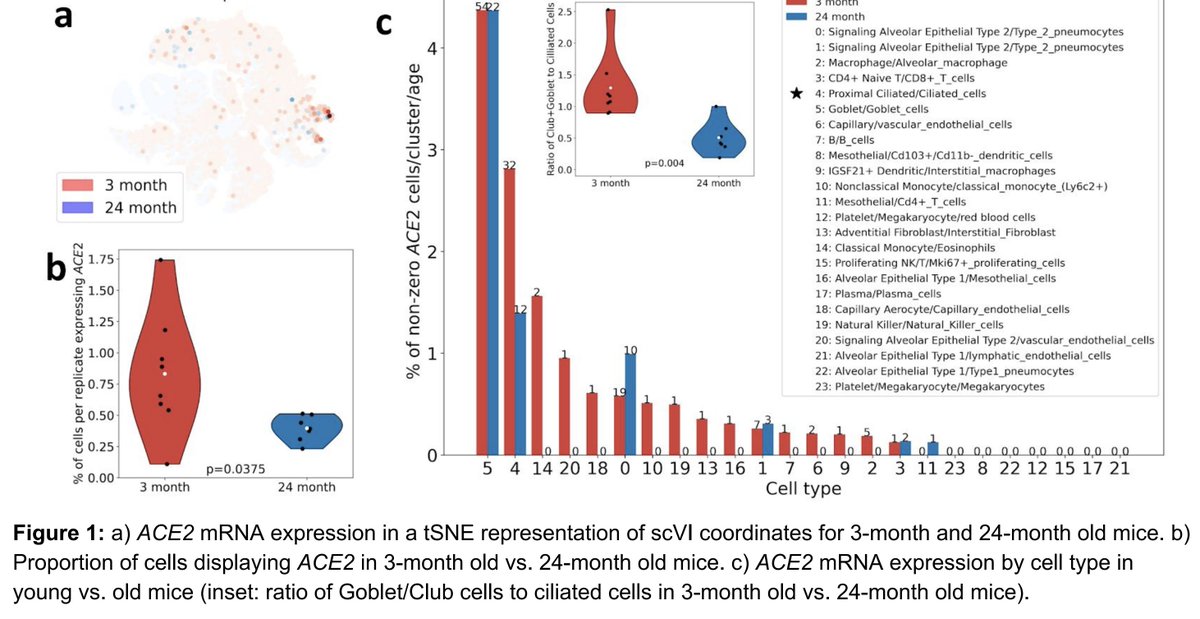
We got a Mac Mini & @pmelsted easily compiled kallisto bustools #scRNAseq on it. Results below:
We got a Mac Mini & @pmelsted easily compiled kallisto bustools #scRNAseq on it. Results below:

Several points:
1. Compilation of code on the M1 ARM architecture was easy for kallisto and bustools because they have few dependencies. In fact we did it before for the ARM Rock64 which is why this time there was no problem with the M1.
1. Compilation of code on the M1 ARM architecture was easy for kallisto and bustools because they have few dependencies. In fact we did it before for the ARM Rock64 which is why this time there was no problem with the M1.
https://twitter.com/lpachter/status/958114838588329984
2. @Apple has done a great job with Rosetta 2. M1 emulating x86 is still faster than previous Macs. And the extra cores are great for running kallisto. macrumors.com/2020/11/15/m1-…
3. The M1 running native code appears to be ~20 faster than the Rosetta 2 emulator. That 5nm fabrication has made a difference! And it turns out that @pmelsted's minimalist engineering has paid off so kallisto and bustools can take advantage of this.
4. I respectfully disagree that nobody wants to run bioinformatics tools on their laptop. It's very convenient at times and many kallisto users work with the software locally.
https://twitter.com/klmr/status/1327596193027862528?s=20
5. @pmelsted and @sinabooeshaghi et al. are wrapping up some improvements to kallisto and bustools with new releases for both tools coming soon. We'll include the M1 binaries with those releases.
• • •
Missing some Tweet in this thread? You can try to
force a refresh