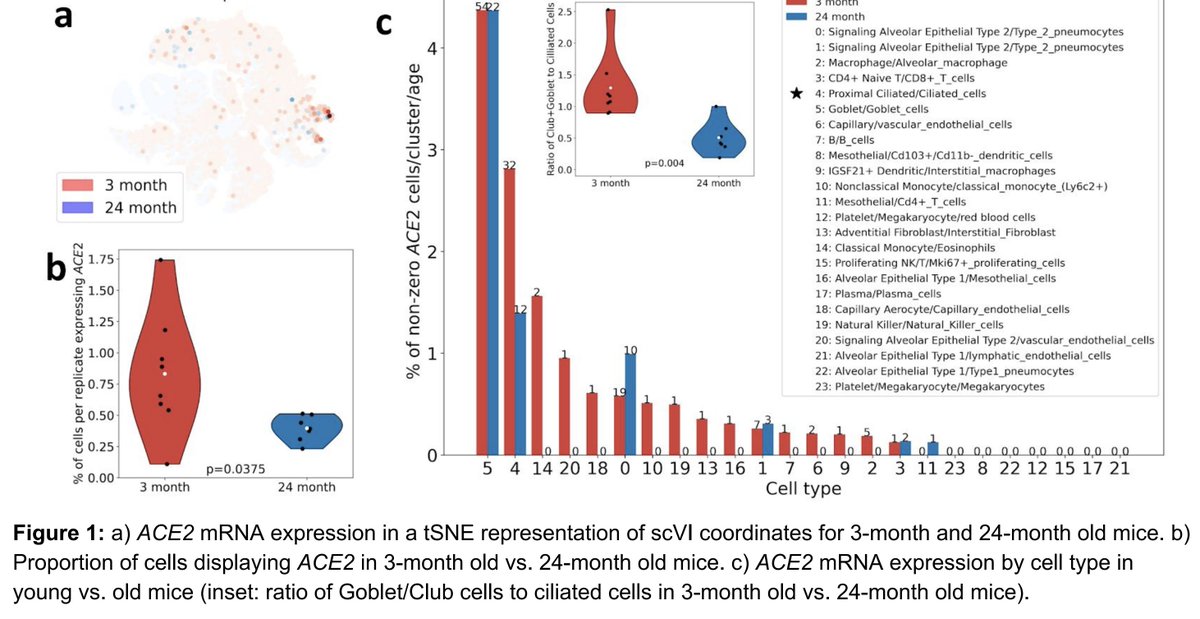
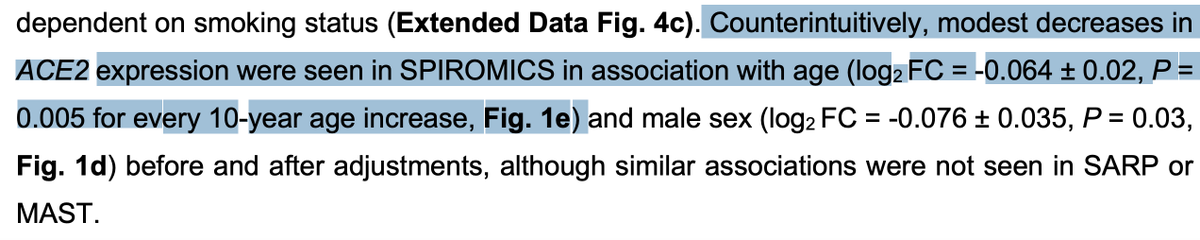
A thread about curated databases in genomics:
The first database I curated by hand was for my Ph.D. thesis. It consisted of a database of 117 orthologous human and mouse genes (this was in the late 90s before either genome was sequenced!). It's still up: cb.csail.mit.edu/cb/crossspecie…
The first database I curated by hand was for my Ph.D. thesis. It consisted of a database of 117 orthologous human and mouse genes (this was in the late 90s before either genome was sequenced!). It's still up: cb.csail.mit.edu/cb/crossspecie…
Compiling this database was hard. It required combing through GENBANK, performing alignments to check for orthology, examine proteins for homology etc. The database was generated for benchmarking a gene prediction tool, but I found that the curation had much more value than that.
The process of compiling the database taught me a ton about the state of gene sequences in GENBANK, challenges in sequence alignment, functional annotation etc. I learned a lot making this database. Also others found it useful in derivative work: korflab.ucdavis.edu/~genis/documen….
Of course my 117 human and mouse genes database is now obsolete. This ends up being the case with most hand curated databases. I think that's ok. The value of engaging in the process is, in my opinion, undervalued. And the databases can be very useful while they last.
One database that is very useful in the single-cell RNA-seq domain right now is this one compiled by @vallens, that we just published:
https://twitter.com/vallens/status/1334904080926466050?s=20
To make this database @vallens didn't just scrape Google Scholar with some script. The detailed information in the different fields required reading the papers. It is a Herculean task, and probably impossible if one started now. @vallens started this not longer after day one.
There is a complementary dataset, not of papers publishing #scRNAseq datasets, but of tools for their analysis made by @_lazappi_: journals.plos.org/ploscompbiol/a…
Together these databases tell an interesting story of a rapidly expanding field, where new datasets are driving tool development and vice versa: 

One of the great things about these databases (unlike mine in the late 90s) is that they can be scraped. The plot above was made using this @GoogleColab notebook today: colab.research.google.com/github/pachter…
Importing the datasets is simple:
https://twitter.com/sinabooeshaghi/status/1331057098386358272?s=20
FYI: Nucleic Acids Research has a database issue every year, and many of the database are valuable, but sadly not all are open and usable in the ways described above.
academic.oup.com/nar/issue/48/D1
academic.oup.com/nar/issue/48/D1
Back to my thesis: I had one hardcopy I kept for the last 20 years. It was stored in a box in our lab and two months ago was destroyed in a flood (thank you 2020!) Then again, half of it consisted of a printout of the entire 117 gene database. It wasn't on anyones reading list...
• • •
Missing some Tweet in this thread? You can try to
force a refresh