Is the new UK variant the same as the new South African variant (501Y.V2)?
No.
They both share the same mutation in spike: N501Y (N->Y at position 501). However, the 2 variants have arisen separately.
1/N
nextstrain.org/groups/neherla…
No.
They both share the same mutation in spike: N501Y (N->Y at position 501). However, the 2 variants have arisen separately.
1/N
nextstrain.org/groups/neherla…

In the tree below, the UK variant is in green at the top. The SA variant is the one with large yellow circles nearer the bottom.
Though it's a bit hard to see in this tree, they are quite distant genetically: they have many mutations they do not share!
2/N
Though it's a bit hard to see in this tree, they are quite distant genetically: they have many mutations they do not share!
2/N

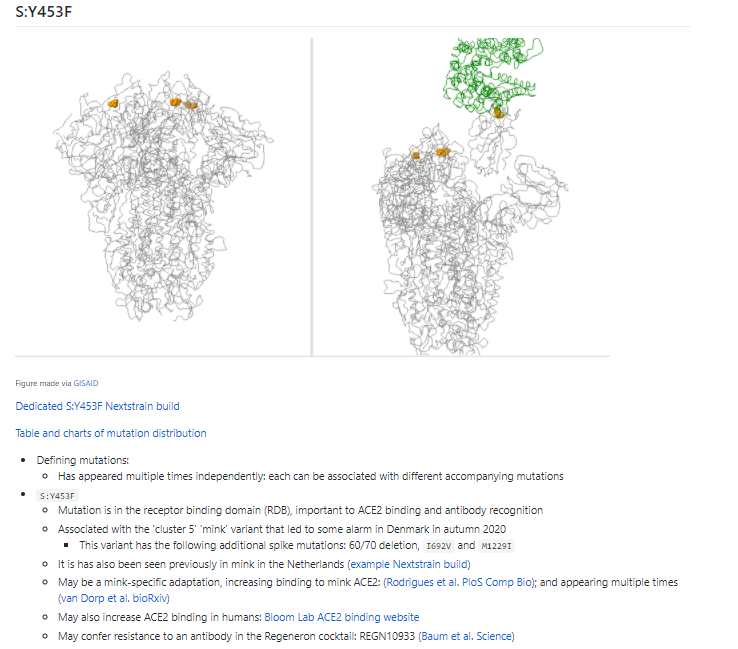
S:N501Y has occurred multiple times around the world. Not all of these have risen & become prominent. So the role of 501Y is probably more complex.
UK variant & SA variant both have many other mutations that are being investigated.
3/N
UK variant & SA variant both have many other mutations that are being investigated.
3/N
Both the UK & SA variants also sit on 'long branches'. This means they have more mutations (different in each) than we would expect. Both also have a higher number of mutations in spike.
Hard to interpret what this means, but another reasons scientists are investigating.
4/N
Hard to interpret what this means, but another reasons scientists are investigating.
4/N
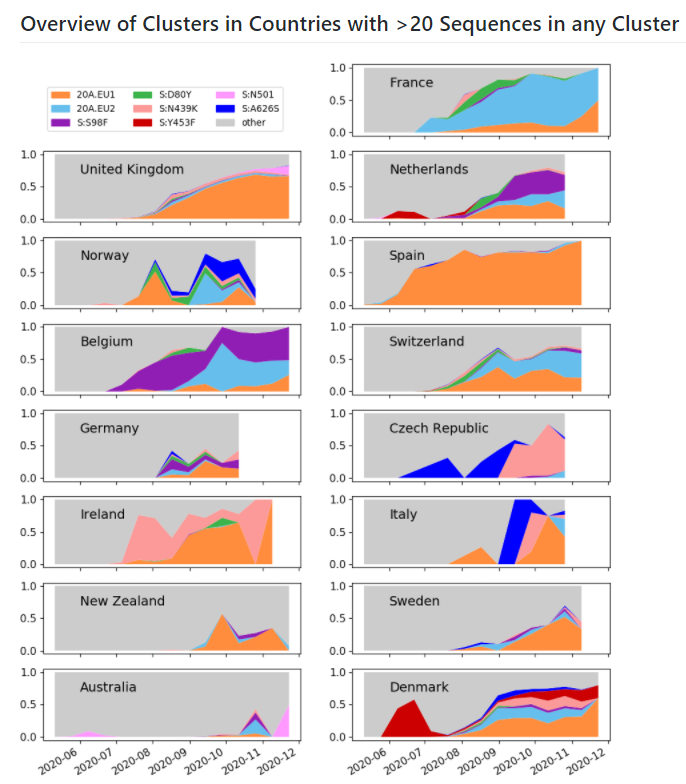
Where have we seen these variants so far?
For the UK variant:
In the UK, we see sequences from England, Wales (a little hard to see) and Scotland. None in Northern Ireland so far.
nextstrain.org/groups/neherla…
5/N
For the UK variant:
In the UK, we see sequences from England, Wales (a little hard to see) and Scotland. None in Northern Ireland so far.
nextstrain.org/groups/neherla…
5/N

For the UK variant:
Outside of the UK, we see very small numbers of sequences in Denmark (a bit hard to see - hidden behind UK circle) & Australia. Their position on the tree indicates that they're likely exports from the UK.
6/N
nextstrain.org/groups/neherla…
Outside of the UK, we see very small numbers of sequences in Denmark (a bit hard to see - hidden behind UK circle) & Australia. Their position on the tree indicates that they're likely exports from the UK.
6/N
nextstrain.org/groups/neherla…

Importantly!:
In Europe, the UK & Denmark are the most regular & prolific sequencers. So the variant could be elsewhere & not detected yet.
In Africa, same is true for South Africa.
More coordinated sequencing efforts regionally & globally would help us monitor variants
8/N
In Europe, the UK & Denmark are the most regular & prolific sequencers. So the variant could be elsewhere & not detected yet.
In Africa, same is true for South Africa.
More coordinated sequencing efforts regionally & globally would help us monitor variants
8/N
I'm re-running the cluster builds now with data from yesterday. I'll post a link when they are done, though I don't expect much will change from the above pictures/links.
9/N
9/N
Are either/both more transmissible? I don't think we know for sure yet, but scientists in the UK seem to believe there are indicators of increased transmission in that variant.
Certainly, we should be extra cautious & each do our part to break transmission chains.
10/N
Certainly, we should be extra cautious & each do our part to break transmission chains.
10/N
Will it impact the vaccine? I don't think we know for sure yet. Hopefully not
The vaccine causes your body to recognise many parts of spike, so a few changes should not mean it stops working. However impact of more changes is less sure - scientists investigating this now.
11/N
The vaccine causes your body to recognise many parts of spike, so a few changes should not mean it stops working. However impact of more changes is less sure - scientists investigating this now.
11/N
Just to add: notifications exploding right now - I'll be missing a lot & can't respond to them all - sorry!
Will try to update this thread when I can, but will probably fall off from individual responses a bit.
12/N
Will try to update this thread when I can, but will probably fall off from individual responses a bit.
12/N
Some vaccine information from @KrutikaKuppalli from the ACIP (Advisory Committee on Immunization Practices - advises the CDC in the USA).
13/N
13/N
https://twitter.com/KrutikaKuppalli/status/1340352974112043009
Very importantly - need to recognise incredible amounts of work on the new variant in South Africa (501Y.V2) by @Tuliodna & @EduanWilkinson at @krisp_news.
And a lot of hard work in the UK on that variant by scientists at @CovidGenomicsUK & SPI-M.
14/N
And a lot of hard work in the UK on that variant by scientists at @CovidGenomicsUK & SPI-M.
14/N
A new run of the S:N501 data is up now - with improved background sequencing to better show how the mutations have arisen independently!
UK variant at top in green. SA variant the blue in the middle - stemming from "20C" label.
15/N
nextstrain.org/groups/neherla…
UK variant at top in green. SA variant the blue in the middle - stemming from "20C" label.
15/N
nextstrain.org/groups/neherla…

• • •
Missing some Tweet in this thread? You can try to
force a refresh