From the latest data available on GISAID this morning, an updated S:N501 #SARSCoV2 build is available.
We now have a clade label for the South African variant (20C.501.V2) & will add one for the UK variant soon.
No major additions to the tree.
1/4
nextstrain.org/groups/neherla…
We now have a clade label for the South African variant (20C.501.V2) & will add one for the UK variant soon.
No major additions to the tree.
1/4
nextstrain.org/groups/neherla…

The latest data identified additional sequences in S:501 mutant variants from the UK, Denmark, Australia, & South Africa, but no additional countries.
(!This table does not distinguish variants & some countries listed haven't seen 501 in months!)
2/4
github.com/emmahodcroft/c…
(!This table does not distinguish variants & some countries listed haven't seen 501 in months!)
2/4
github.com/emmahodcroft/c…
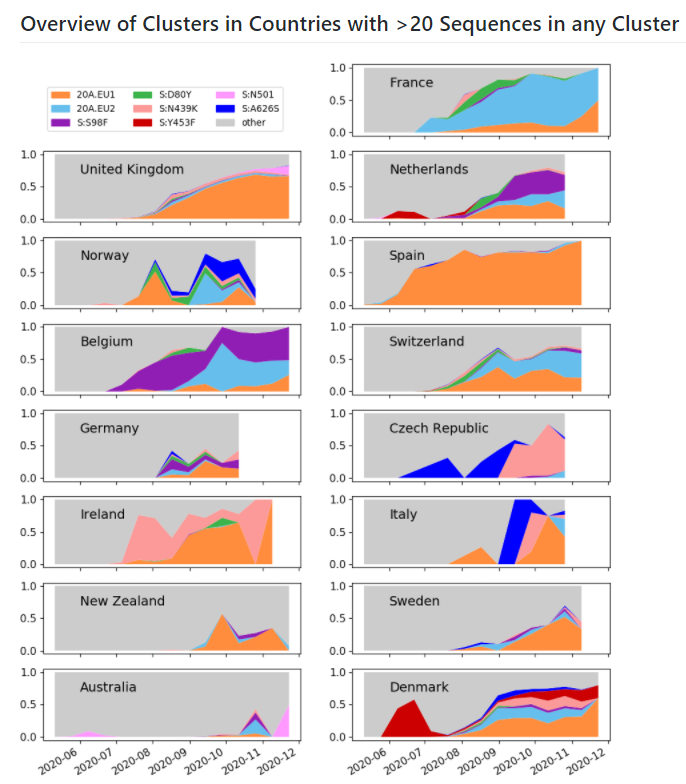
From the plot of variants frequency in sequences, per country, we can see that the 501Y variant (pink) is expanding in the UK (the last data point represents incomplete data & will likely change in future).
3/4
github.com/emmahodcroft/c…
3/4
github.com/emmahodcroft/c…

Seeing no new countries in sequences isn't at odds with media reports: our analysis is based only on sequences which are made available to scientists through GISAID. Countries can analyse their own sequences faster, so they may be detecting cases we do not have samples for.
4/4
4/4
For more information on the new UK and SA variants, check out my thread from yesterday:
https://twitter.com/firefoxx66/status/1340359989395861506
• • •
Missing some Tweet in this thread? You can try to
force a refresh