🎉Happy New Year!🎉
To kick off 2021, here's an update on:
- S:N501 variants (501Y.V1/V2) with data from 31 Dec 2020
- 69/70del variants
- 20A.EU1 variant (most prevalent variant in Europe)
(Here are some fireworks that look a bit like phylogenies! 🎆)
1/23
To kick off 2021, here's an update on:
- S:N501 variants (501Y.V1/V2) with data from 31 Dec 2020
- 69/70del variants
- 20A.EU1 variant (most prevalent variant in Europe)
(Here are some fireworks that look a bit like phylogenies! 🎆)
1/23

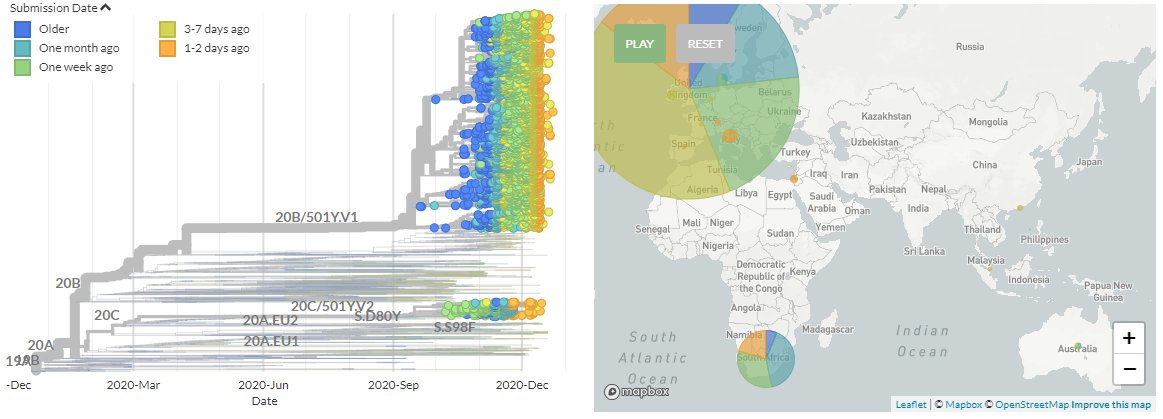
The latest sequences from 31 Dec show new sequences in the S:N501Y.V1 variant (originating from SE England) from Netherlands, Denmark, Australia, Italy, Canada, the USA, Germany, Switzerland, & India.
2/23
S:N501 link
nextstrain.org/groups/neherla…
Image link
nextstrain.org/groups/neherla…
2/23
S:N501 link
nextstrain.org/groups/neherla…
Image link
nextstrain.org/groups/neherla…

Canada, Germany, & Switzerland all have sequences that fall in 501Y.V1 for the first time, scattered across the tree.
3/23
3/23
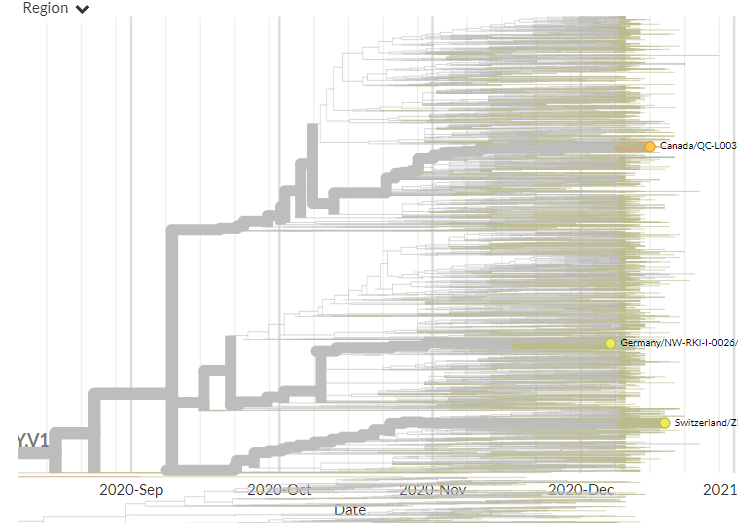
The USA also has its first 2 sequences that fall in 501Y.V1, from Colorado & California. India also had its first 3 sequences, also from different regions.
In both cases, the sequences are not connected, indicating separate introductions.
4/23

In both cases, the sequences are not connected, indicating separate introductions.
4/23
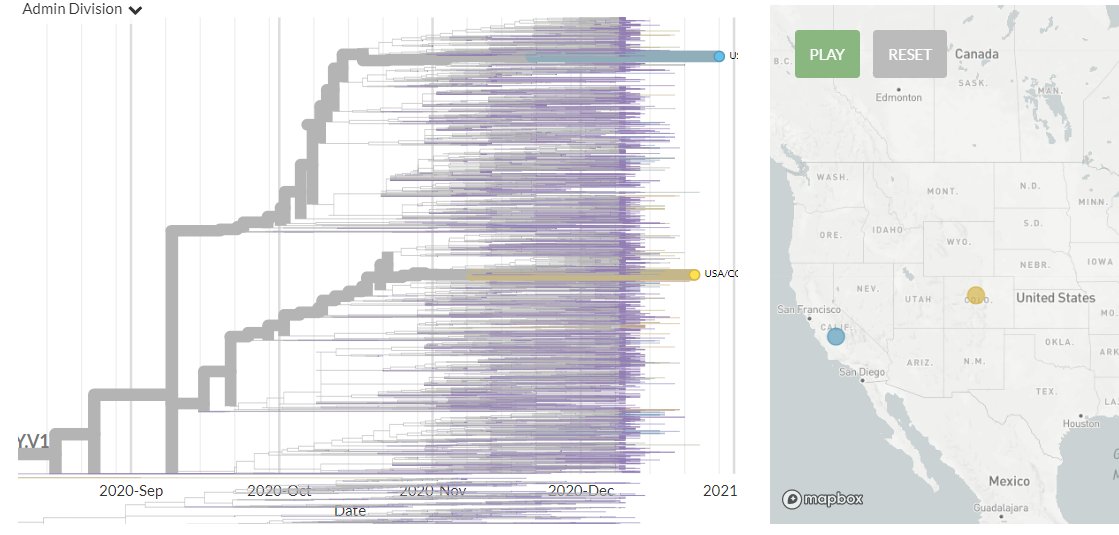

In countries with previous sequences:
6 new sequences from Netherlands, in orange/yellow (green are older).
1 is clearly separate - zooming in (& in divergence view, coloured by country) it is harder to determine the relationship between the others.
5/23

6 new sequences from Netherlands, in orange/yellow (green are older).
1 is clearly separate - zooming in (& in divergence view, coloured by country) it is harder to determine the relationship between the others.
5/23


Denmark has 24 new sequences, shown in orange.
2 identical sequence at the top are separate introduction(s?) than the larger cluster below.
The other sequences form a tight cluster very indicative of onward transmission.
6/23

2 identical sequence at the top are separate introduction(s?) than the larger cluster below.
The other sequences form a tight cluster very indicative of onward transmission.
6/23


1 new Australian sequence (the large orange dot) is close to an older sequence (large green).
From the phylogeny alone, it's hard to say if these are connected or separate, as the sequences are identical. (Vertical order is not meaningful for identical sequences!)
7/23
From the phylogeny alone, it's hard to say if these are connected or separate, as the sequences are identical. (Vertical order is not meaningful for identical sequences!)
7/23

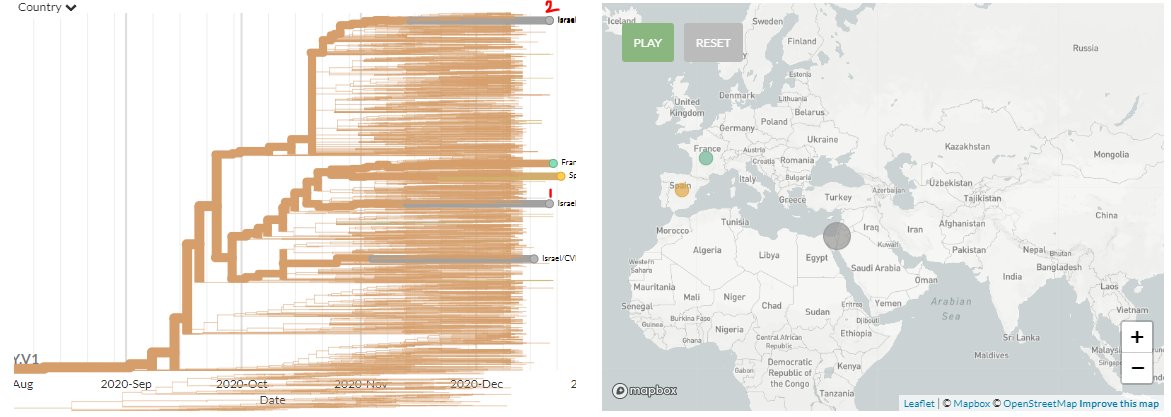
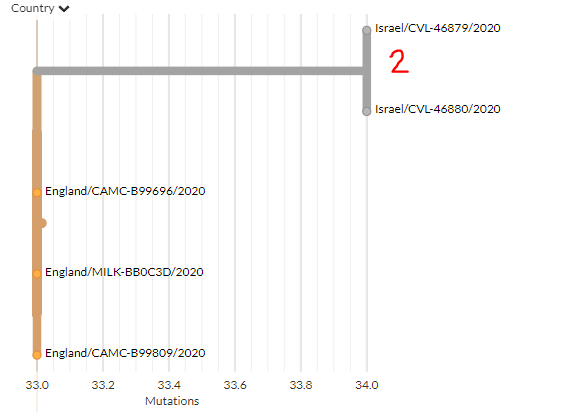
Finally, there are 2 new sequences from Italy (orange).
1 (top) seems to be a separate introduction. The other (zoomed view) may be related to an earlier Italian sequence as they're separated by 1 mutation, but we cannot say for sure.
8/23

1 (top) seems to be a separate introduction. The other (zoomed view) may be related to an earlier Italian sequence as they're separated by 1 mutation, but we cannot say for sure.
8/23


Now: 69/70del variants. Why do we care about 69/70del? Well, this deletion is found in 501Y.V1, but also in other major variants like S:Y453F & S:N439K.
Read/see more in the 69del section of my 'SARS-CoVariants' tracking page!
9/23
github.com/emmahodcroft/c…
Read/see more in the 69del section of my 'SARS-CoVariants' tracking page!
9/23
github.com/emmahodcroft/c…

In a new Virological post @bblarsen1
& @MichaelWorobey identify a novel 69/70del lineage in the USA:
virological.org/t/identificati…
10/23
& @MichaelWorobey identify a novel 69/70del lineage in the USA:
virological.org/t/identificati…
10/23
In the focal S:H69- build, we can see this cluster below.
Importantly, in this build the cluster appears to be close to 501Y.V2, but this is a focal build so this connection must be interpreted cautiously!
11/23
nextstrain.org/groups/neherla…
Importantly, in this build the cluster appears to be close to 501Y.V2, but this is a focal build so this connection must be interpreted cautiously!
11/23
nextstrain.org/groups/neherla…

We can also see the cluster in our soon-to-go-live-today @nextstrain North American build (subsampled, in grey).
(In the build live at 14.30CET 1 Jan the 'cluster' is only 1 sequence! We'll push the new one later today!)
12/23
(In the build live at 14.30CET 1 Jan the 'cluster' is only 1 sequence! We'll push the new one later today!)
12/23

Notably we do see 69/70del clusters pop up often (as well as individuals/pairs with the deletion) - here's just a few:
18 sequences from the UK, Denmark, Germany & France.
13/23
nextstrain.org/groups/neherla…
18 sequences from the UK, Denmark, Germany & France.
13/23
nextstrain.org/groups/neherla…

A small cluster of 4 sequences from England, Germany, Australia.
nextstrain.org/groups/neherla…
A small cluster of 6 sequences from England.
nextstrain.org/groups/neherla…
14/23

nextstrain.org/groups/neherla…
A small cluster of 6 sequences from England.
nextstrain.org/groups/neherla…
14/23


A cluster of 8 sequences from the Ivory Coast and Burkina Faso.
15/23
nextstrain.org/groups/neherla…
15/23
nextstrain.org/groups/neherla…

We can also see 69/70del popping up inside of variants other than the recognised 501Y.V1, 453, & 439:
We can see ~10 sequences in 20A.EU2 (S:477) from France with 69/70del. Gaps aren't used in the phylogeny building, so they don't cluster here.
16/23
nextstrain.org/groups/neherla…
We can see ~10 sequences in 20A.EU2 (S:477) from France with 69/70del. Gaps aren't used in the phylogeny building, so they don't cluster here.
16/23
nextstrain.org/groups/neherla…
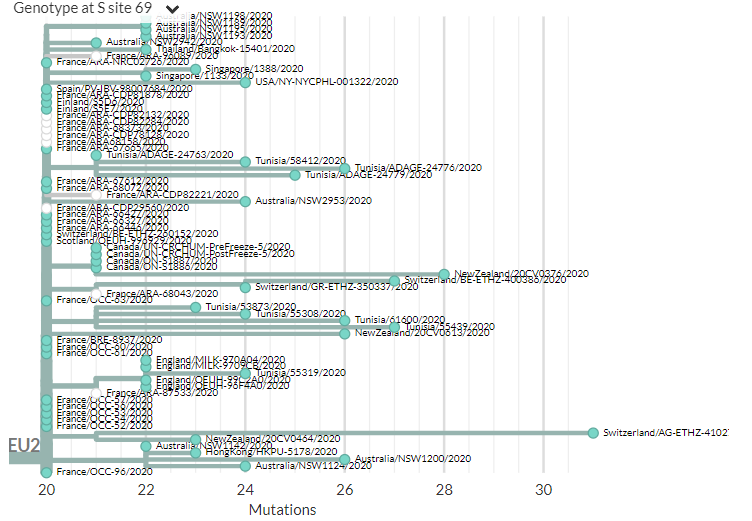
Interestingly, @richardneher has queried all the 69/70 deletions in SARS-CoV-2 sequences (all across the tree), and they all occur with the same nucleotide pattern, implying this is a good way to get this deletion, for one reason or another.
17/23
17/23
https://twitter.com/richardneher/status/1345003715611267072
Finally, a brief note on 20A.EU1.
This is the variant that expanded in Spain before spreading across Europe over the summer (you can read more in our preprint below). It is the most prevalent variant in Europe, containing ~64,000 sequences.
18/23
medrxiv.org/content/10.110…
This is the variant that expanded in Spain before spreading across Europe over the summer (you can read more in our preprint below). It is the most prevalent variant in Europe, containing ~64,000 sequences.
18/23
medrxiv.org/content/10.110…

We have no evidence that the mutations in 20A.EU1 (notably S:A222V) make it more transmissible - we think its spread was mostly due to summer travel.
But long after summer travel has ended, it persists (in orange below) - & continues to spread...
19/23
github.com/emmahodcroft/c…
But long after summer travel has ended, it persists (in orange below) - & continues to spread...
19/23
github.com/emmahodcroft/c…

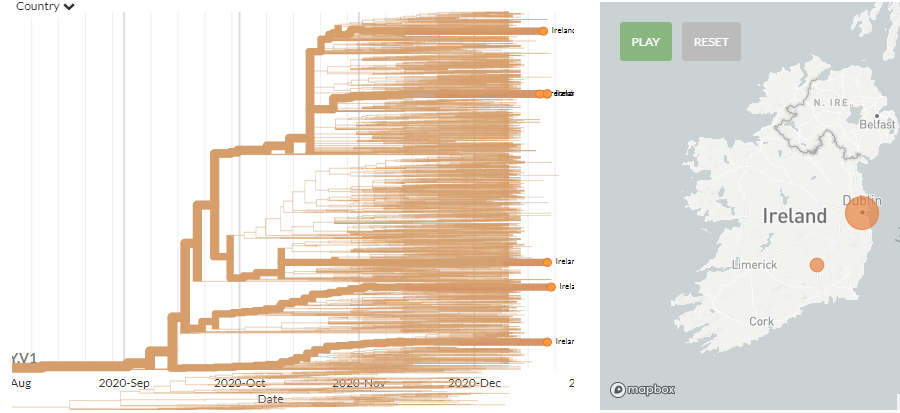
In the last 2 weeks we've seen the first appearance of sequences in 20A.EU1 in the USA, Thailand, Malaysia, Peru, Finland, Israel, & South Korea - all sampled in autumn.
20/23
github.com/emmahodcroft/c…
20/23
github.com/emmahodcroft/c…
This is a variant with no evidence of increased transmissibility, during a time when global travel is severely depressed.
20A.EU1's movement across Europe & the world show how effectively & efficiently #SARSCoV2 #COVID19 can spread.
21/23
nextstrain.org/groups/neherla…
20A.EU1's movement across Europe & the world show how effectively & efficiently #SARSCoV2 #COVID19 can spread.
21/23
nextstrain.org/groups/neherla…

Our #SARSCoV2 strategies must take this into account: getting cases down in your country won't last if they are being imported from elsewhere.
And it reminds us how vital & informative it is to track the virus via phylogenetics - only possible with good sequencing.
22/23
And it reminds us how vital & informative it is to track the virus via phylogenetics - only possible with good sequencing.
22/23
2020 has often been a stressful, sad, lonely, desperate, scary year.
But it has also been one where we've seen what science can accomplish when resources are made available & scientists work together.
With all we leave behind in 2020, let's carry that into 2021!
23/23
But it has also been one where we've seen what science can accomplish when resources are made available & scientists work together.
With all we leave behind in 2020, let's carry that into 2021!
23/23

Apologies, forgot to add: The 'reduced UK' S:N501 focal build is also updated, for those who find this useful. As before, *do not use this build to look at connections between countries* - use the 'full' N501 build for this.
nextstrain.org/groups/neherla…
nextstrain.org/groups/neherla…

• • •
Missing some Tweet in this thread? You can try to
force a refresh