Proprietary prediction models are widely implemented in health care. Let’s talk about why they exist and if we can (or should) move away from them.
Let's start with a poll. I'll return to this soon with a story about the slow death of an open model.
Why are they used at all?
Let's start with a poll. I'll return to this soon with a story about the slow death of an open model.
Why are they used at all?
All are partially true (will explain) but C is right. Proprietary models are used bc EHRs suffer from a last-mile problem. While scientists debate whether models should even be made available (nature.com/articles/s4158…) the truth is that we don’t have many ways to implement models.
From a technical standpoint, formats *do exist* to facilitate sharing of models (pubmed.ncbi.nlm.nih.gov/33079583/ & nature.com/articles/s4158…) but this is moot bc those formats are (mostly) not supported inside the EHR.
(Check out the @nature letter to @Google informing them about Colab)
(Check out the @nature letter to @Google informing them about Colab)
So, the reason that hundreds of US hospitals use proprietary prediction models today has to do with the fact that these are EHR vendor-developed models and are thus easiest to implement in the EHR itself.
But I said all of the answers were partially right. How is that possible?
But I said all of the answers were partially right. How is that possible?
It's worth considering what a proprietary model actually is.
Which definition best captures the characteristics of a proprietary model?
Is it a model whose...
...variables are not known?
...form/coefficients are not known?
...performance is not known
...can't be used w/o $?
Which definition best captures the characteristics of a proprietary model?
Is it a model whose...
...variables are not known?
...form/coefficients are not known?
...performance is not known
...can't be used w/o $?
D is most correct but others can be true. EHR vendors usually provide information on which variables (+/- actual coefficients) and info on model validation performed at other institutions. If implemented, vendors will even calculate local performance.
But can you trust it?
But can you trust it?
Having read a dozen+ proprietary model briefs, the quality of validation (and assumptions) are highly variable. Also, some vendors are more aggressive than others re subjecting models to peer review. But vendors do privately share validation info with hospitals.
Now for a story.
Now for a story.
Let's talk about APACHE, a series of models that help ICUs assess whether their ICU mortality is better or worse than expected based on patient severity. The story comes straight from its developer (Dr. William Knaus).
1. jamanetwork.com/journals/jamas…
2. mdcalc.com/apache-ii-score

1. jamanetwork.com/journals/jamas…
2. mdcalc.com/apache-ii-score

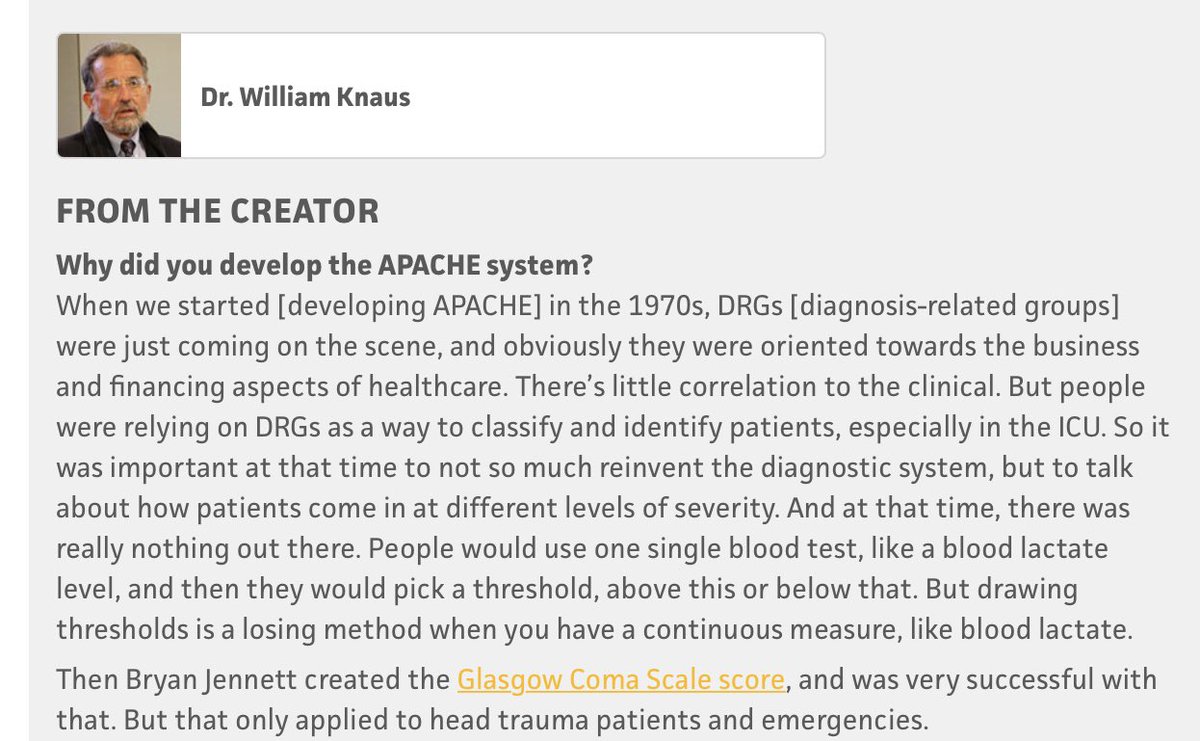
APACHE was invented in 1978 in response to the unexpected death of a young patient. APACHE I was developed in 582 pts and published in 1981. It was tested in France, Spain, and Finland. It was somewhat complex (requiring 33 physiologic measurements), which limited adoption. 

APACHE II reduced the complexity of APACHE (used only 12 physiologic measurements) and adoption was rapid.
The system worked well and was an open model.
You can try it out here: mdcalc.com/apache-ii-score
... then the problems began.
The system worked well and was an open model.
You can try it out here: mdcalc.com/apache-ii-score
... then the problems began.

The 1st problem was that carrying out this international effort to standardize quality measurement was expensive. A company was formed and $ was raised from venture capital.
The 2nd problem was that poor performers doubted the accuracy of APACHE II.
Solution? APACHE III.
The 2nd problem was that poor performers doubted the accuracy of APACHE II.
Solution? APACHE III.

APACHE III improved the AUC from 0.86 (APACHE II) to 0.90. It also addressed issues specific to surgery, trauma, comatose status, etc.
But unlike APACHE II, APACHE III was proprietary.
And it cost money, which led to an investigation re: misuse of funds.
But unlike APACHE II, APACHE III was proprietary.
And it cost money, which led to an investigation re: misuse of funds.

Many ICU physicians were also not pleased with the prospect of paying for the score.
When told about the cost required to run the company and calculate the scores, Dr. Knauss was told to go "get more grants," which wasn't really an option.
...then APACHE got bought by Cerner.
When told about the cost required to run the company and calculate the scores, Dr. Knauss was told to go "get more grants," which wasn't really an option.
...then APACHE got bought by Cerner.

Cerner is one of the two largest EHR vendors in ths US (alongside Epic). Since ICUs generally found APACHE III useful but didn't want to pay for it, it seems ideal that they got bailed out by Cerner, right?
Kind of like how Microsoft bailed out GitHub?
...so what did Cerner do?
Kind of like how Microsoft bailed out GitHub?
...so what did Cerner do?

Cerner unveiled.... *drumroll*
APACHE IV!
Features:
- better calibrated than APACHE III
- more complex than APACHE II/III
"Also we recommend APACHE II no longer be used..."
So how complex was it?

APACHE IV!
Features:
- better calibrated than APACHE III
- more complex than APACHE II/III
"Also we recommend APACHE II no longer be used..."
So how complex was it?

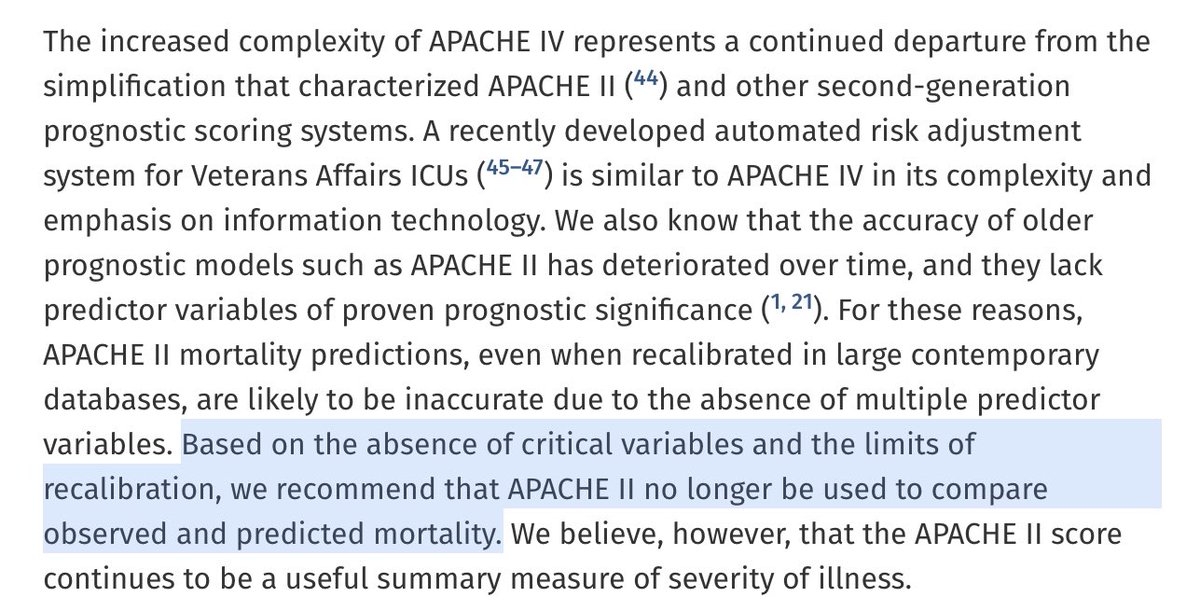
APACHE IV is so complex that centers often perform manual chart validation to confirm that the elements going into the model are accurate.
Source: journals.lww.com/ccmjournal/Ful…
Also, conveniently, APACHE IV isn't integrated with the Epic EHR (hmm wonder why?)
Source: journals.lww.com/ccmjournal/Ful…
Also, conveniently, APACHE IV isn't integrated with the Epic EHR (hmm wonder why?)

Meanwhile, in non-proprietary land, the SAPS-3 model tried to resurrect the simplicity of APACHE II -> simple, but not as good (ncbi.nlm.nih.gov/pmc/articles/P…)
Also, Epic introduced a proprietary ICU mortality prediction model that appears to emulate APACHE IV and is easy to integrate.
Also, Epic introduced a proprietary ICU mortality prediction model that appears to emulate APACHE IV and is easy to integrate.

So which would you use?
- a complex proprietary model owned by Cerner (APACHE IV)
- a simple prediction model (MPM-3) also owned by Cerner
- the proprietary Epic ICU mortality model
- the non-proprietary SAPS-3 (performed worse in an independent validation)
- outdated APACHE II
- a complex proprietary model owned by Cerner (APACHE IV)
- a simple prediction model (MPM-3) also owned by Cerner
- the proprietary Epic ICU mortality model
- the non-proprietary SAPS-3 (performed worse in an independent validation)
- outdated APACHE II
Dr. Knauss, inventor of APACHE, has this to say in a footnote on the MDCalc page for APACHE II (mdcalc.com/apache-ii-score)
"In retrospect, if we had known the future was going to be as limited in the development of health IT, I think we would've said, let's stay with APACHE II."
"In retrospect, if we had known the future was going to be as limited in the development of health IT, I think we would've said, let's stay with APACHE II."

If you work in an ICU, I'd love to know:
What does your ICU actually use to measure how well it is doing in terms of expected vs. observed mortality?
What does your ICU actually use to measure how well it is doing in terms of expected vs. observed mortality?
So what's the moral of the story?
Proprietary models are here to stay (for now), but we need to urgently adopt mechanisms to disseminate and operationalize open-source models in the EHR. This is available in some EHRs but not all. And it's competely different for each EHR.
Proprietary models are here to stay (for now), but we need to urgently adopt mechanisms to disseminate and operationalize open-source models in the EHR. This is available in some EHRs but not all. And it's competely different for each EHR.
Closing thoughts (1/2): We can d/l our patient records today bc of the Blue Button and @myopennotes initiatives.
@calonghurst @drnigam proposed a "Green Button" initiative to get aggregate patient statistics at the bedside (healthaffairs.org/doi/full/10.13…), an important next step.
@calonghurst @drnigam proposed a "Green Button" initiative to get aggregate patient statistics at the bedside (healthaffairs.org/doi/full/10.13…), an important next step.
Closing thoughts (2/2): I'll go further and say that we need an OpenModel initiative that allows prediction models to interface in a constent manner with all EHRs. Not just PMML (model format) but communication stds.
Without it, the future consists mostly of proprietary models.
Without it, the future consists mostly of proprietary models.
• • •
Missing some Tweet in this thread? You can try to
force a refresh