Did DeepMind do the impossible? What can we learn from this? A step-by-step guide.
nature.com/articles/s4158…

cjasn.asnjournals.org/content/11/11/… and
insights.ovid.com/crossref?an=00…
The 2016 @CJASN paper used logistic regression and 2018 paper used GBMs.

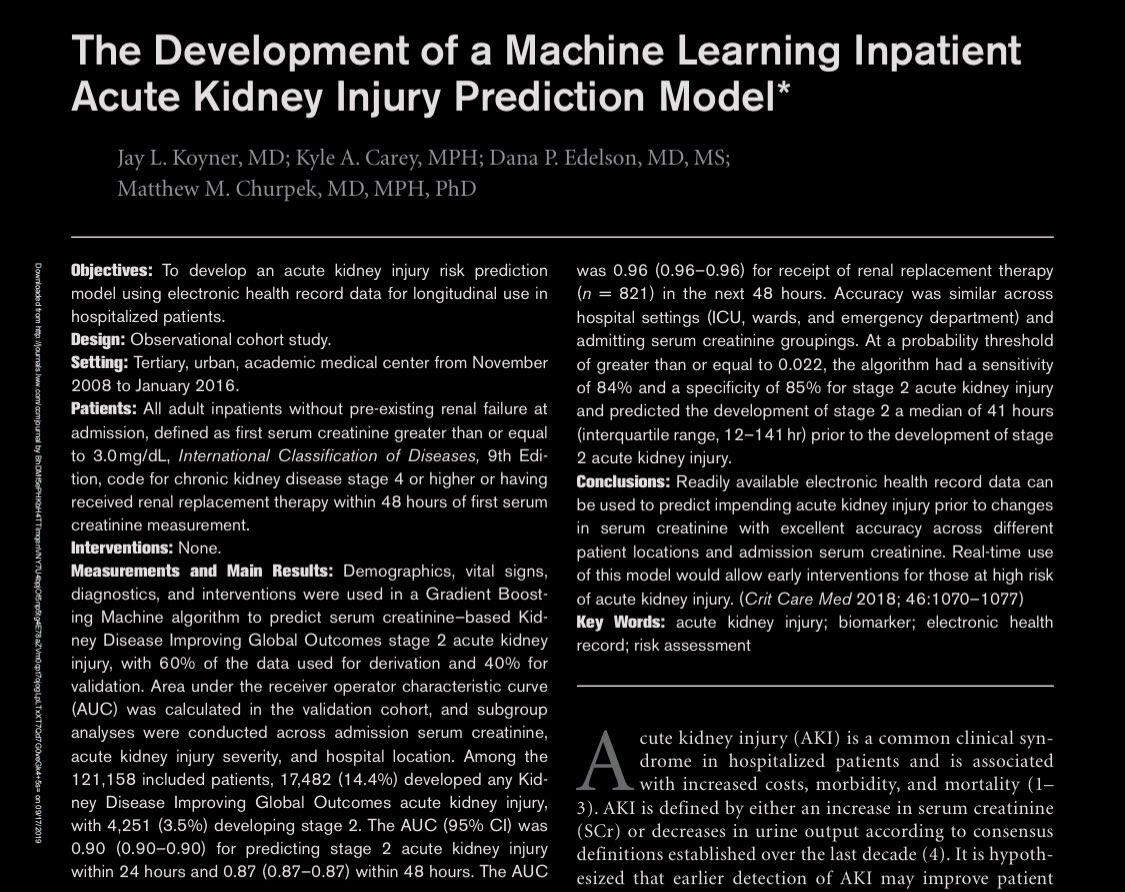
What the heck is that?
- selected interval wasn’t included in paper (“eg 12 hrs”)
- outcome window: 24 hrs
- lookback period was undefined
- missing values imputed by carrying forward
- Cr slope was included as predictor but no other slopes
- Cont variables modeled with splines
Keeping in mind Koyner’s 2018 AUC of 0.73 for 48 hr AKI using GBM, the DeepMind team’s AUC for 48 hr AKI is....
... drumroll ... 0.89! Wait what? How is that possible?
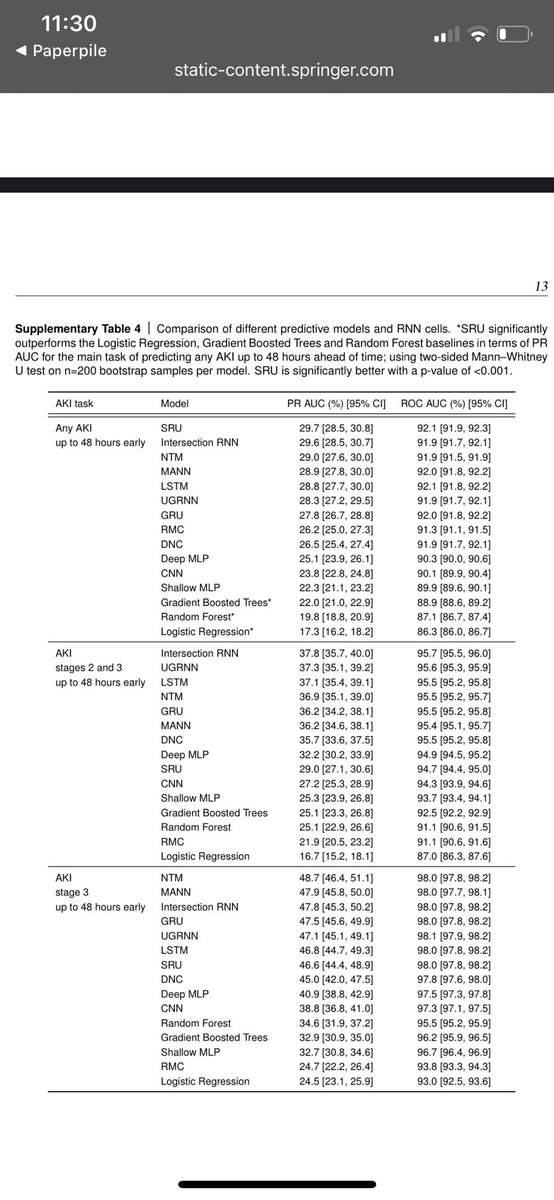
So yes the headline is that an RNN achieved an AUC of 0.92, but the subheadline here is that other approaches (including the same GBM approach used in Koyner’s 2018 paper) got close. Why?
- for each interval, many summary stats for each predictor were included (count, mean, median, sd, min, max)
- lagged and differenced predictors were included
The RNN trained on a 48 hour outcome window but directly modeled 6 hour “steps” along the way.
RNNs process data in batches and learn a state representation (coefficients) that serve as additional input to the next batch.
Ok I think I’ll end there. Let’s discuss this paper. What did you find interesting?
Creatinine is an imperfect gold standard for AKI. It’s the standard of care at the moment but measurement error in the outcome is never a good thing.
ncbi.nlm.nih.gov/pmc/articles/P…
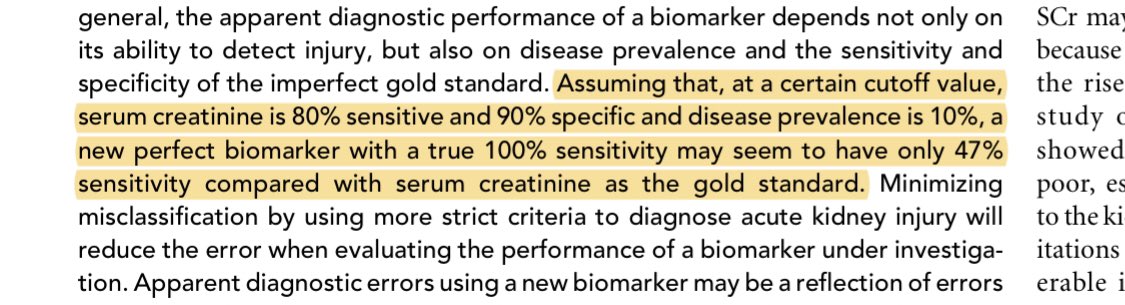
... which is the same way I read this paper because somehow the methods section is AFTER the references? What kind of sorcery is this?