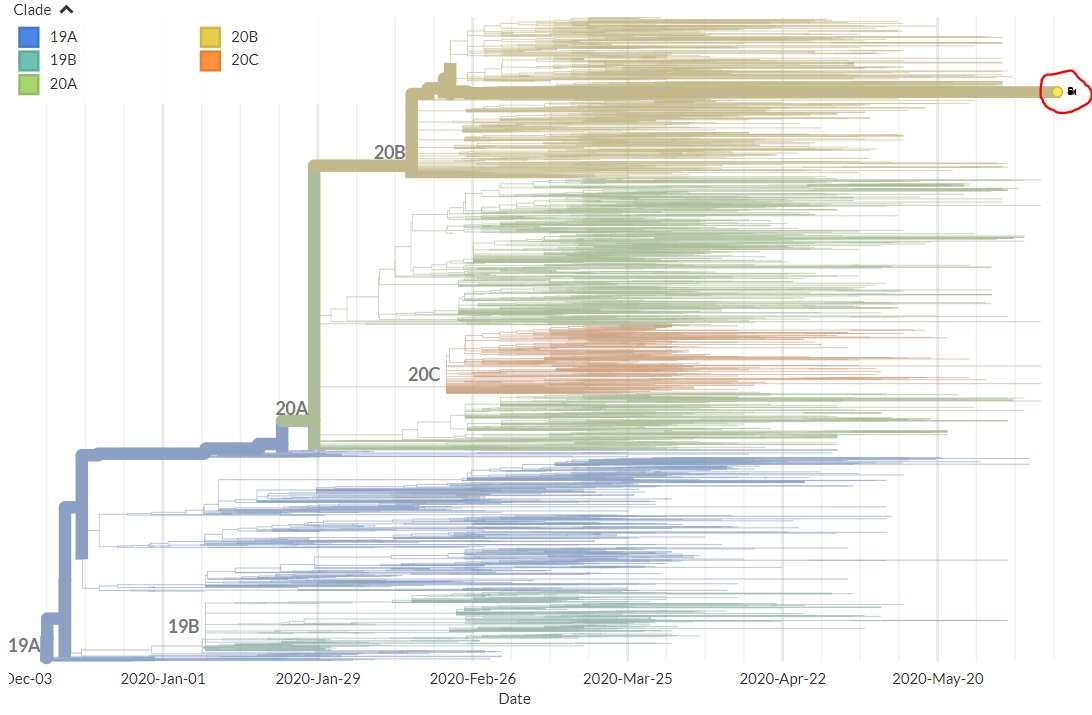
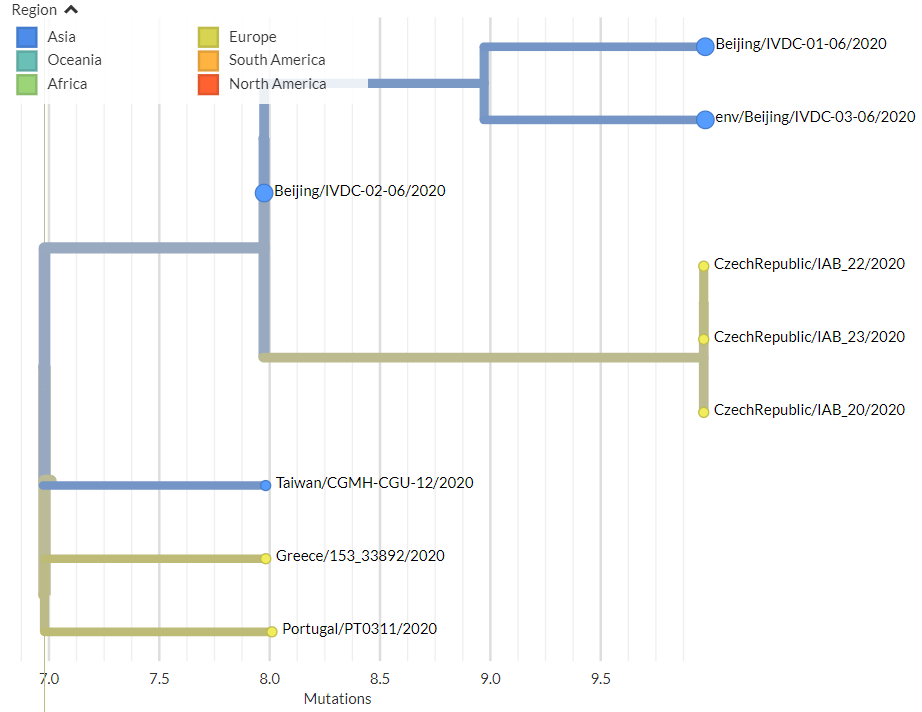
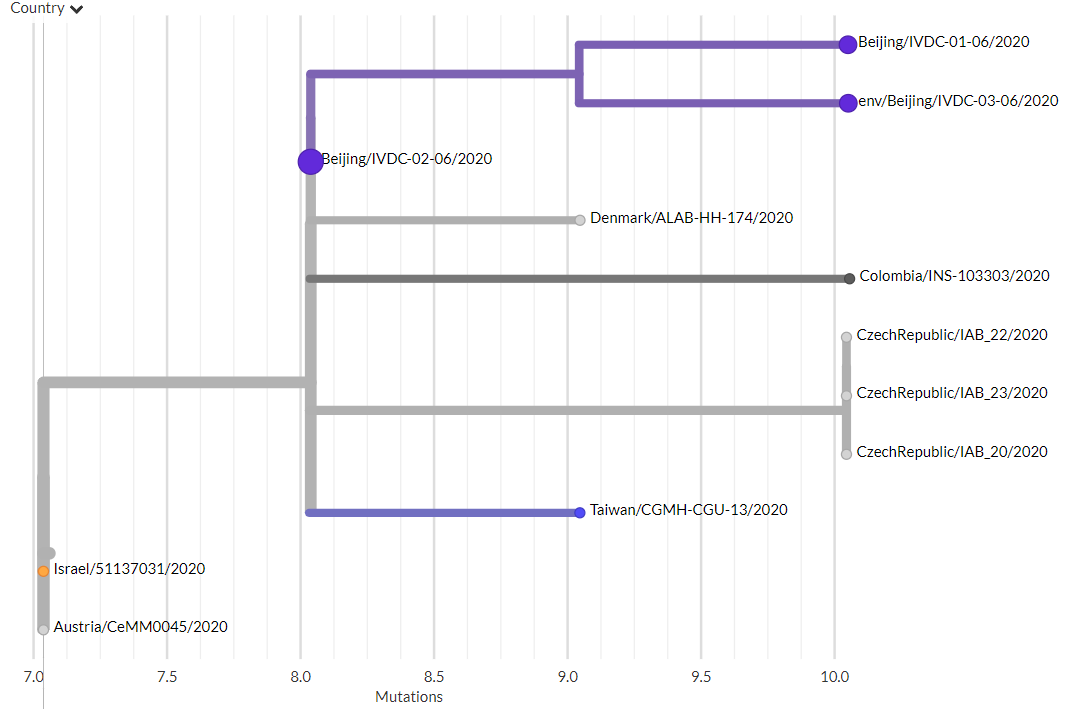
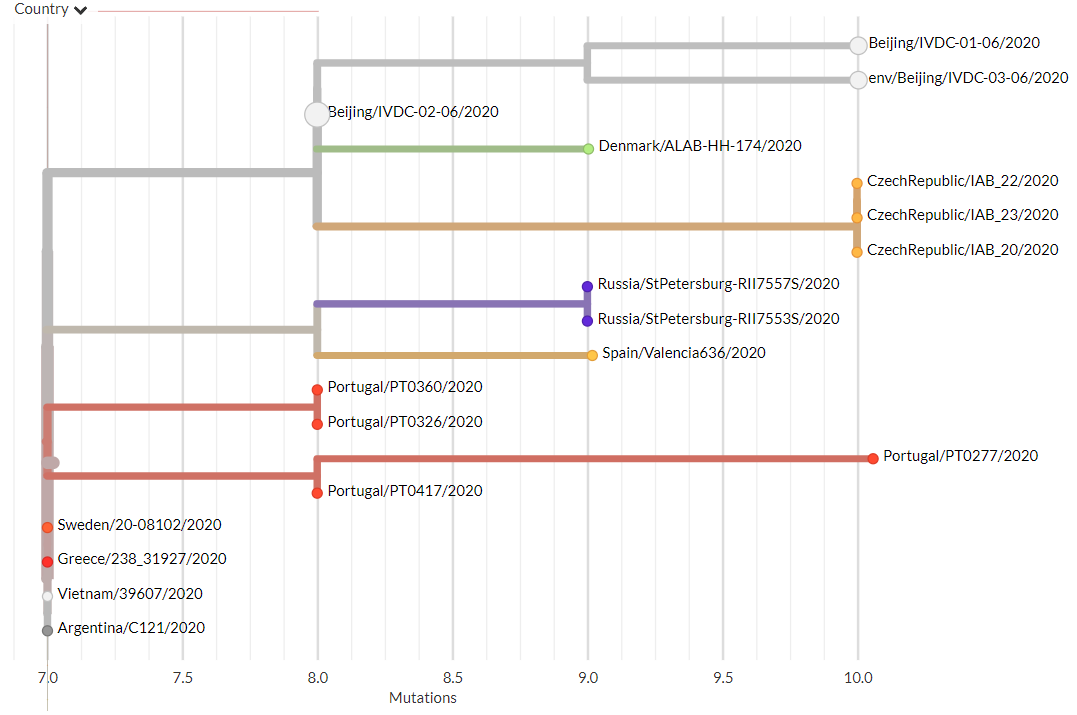
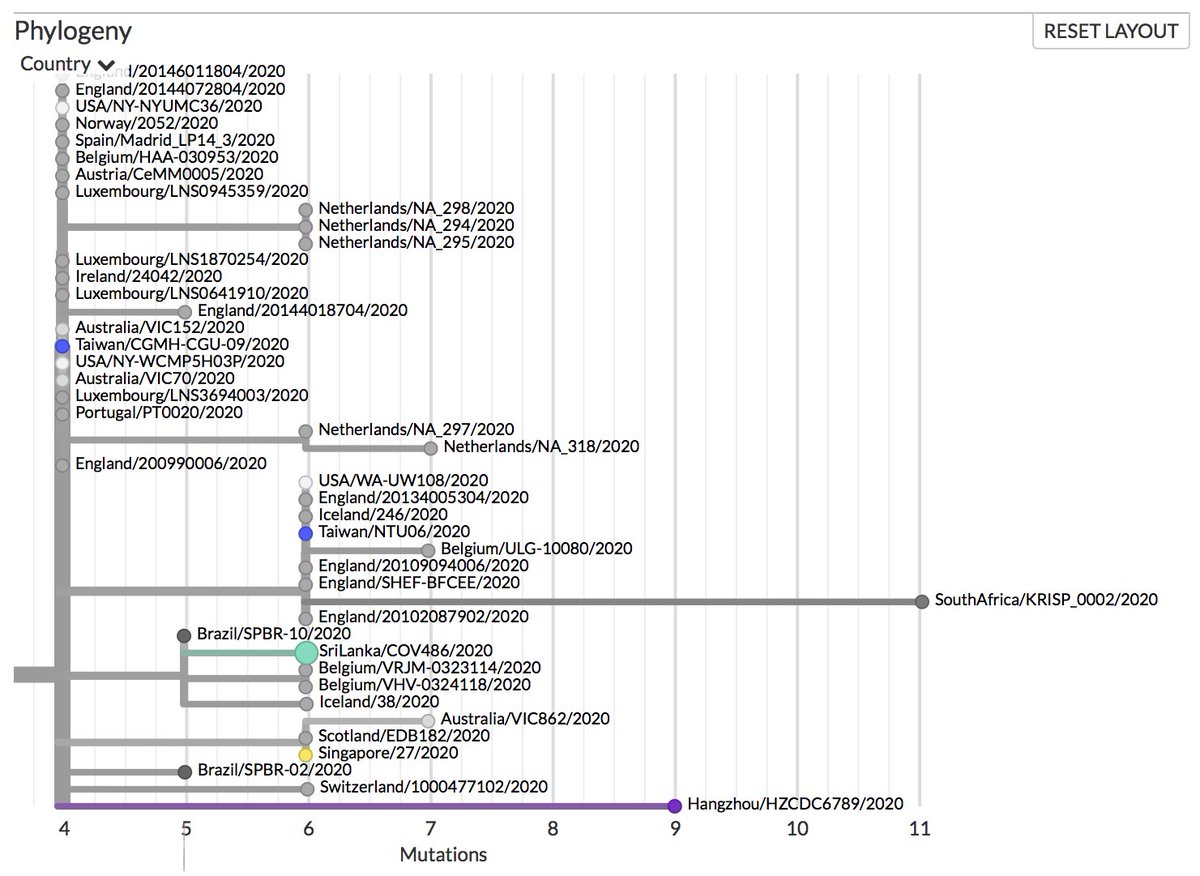
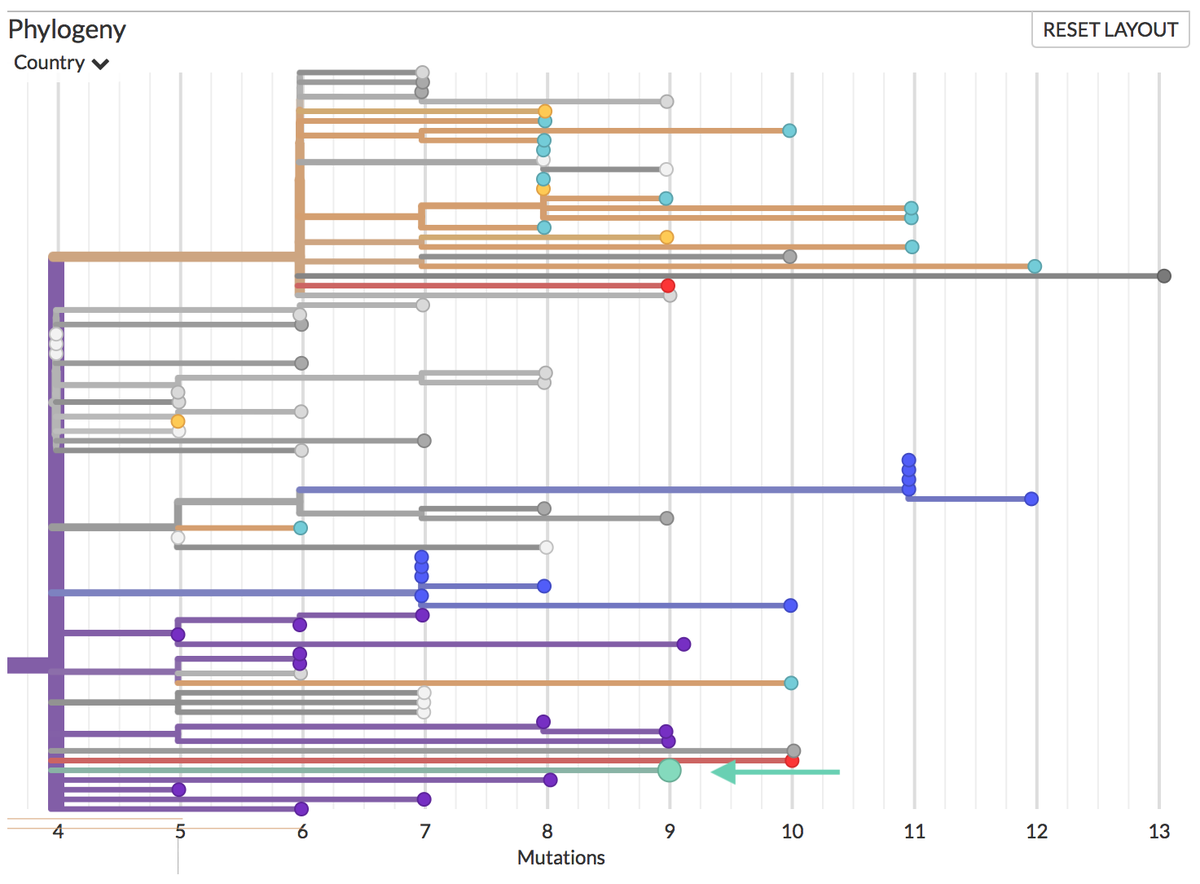
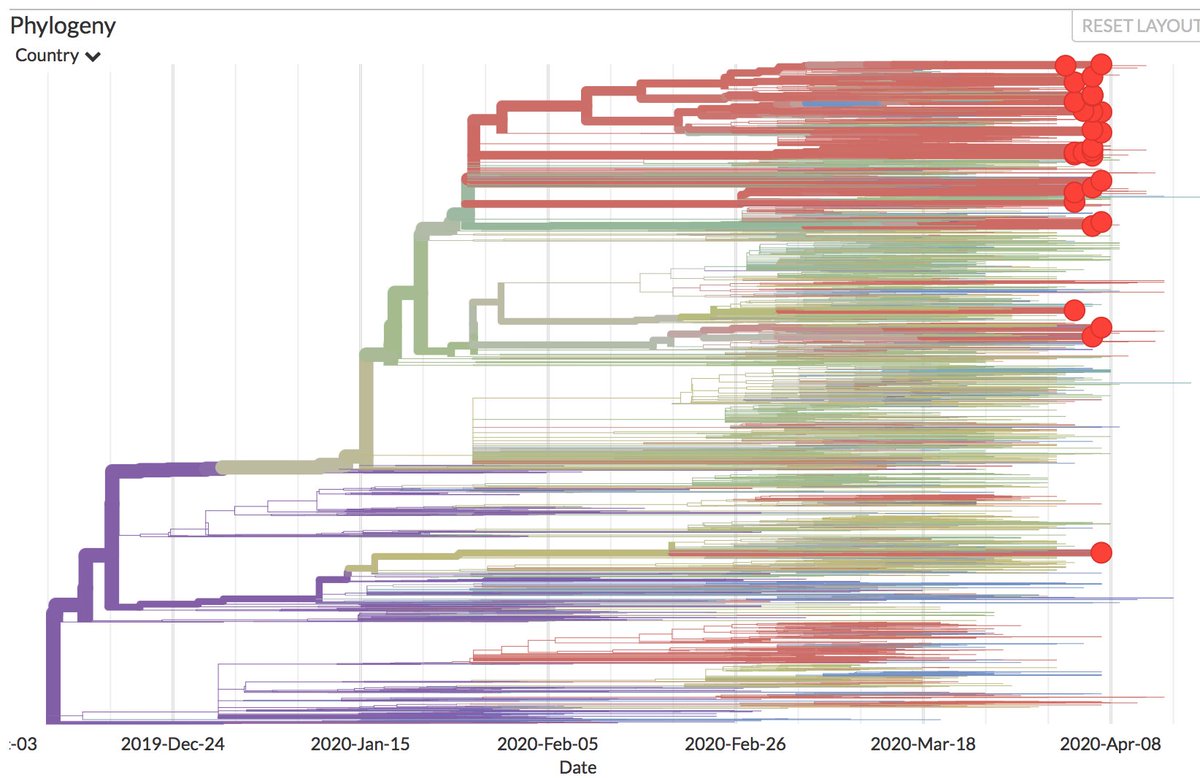
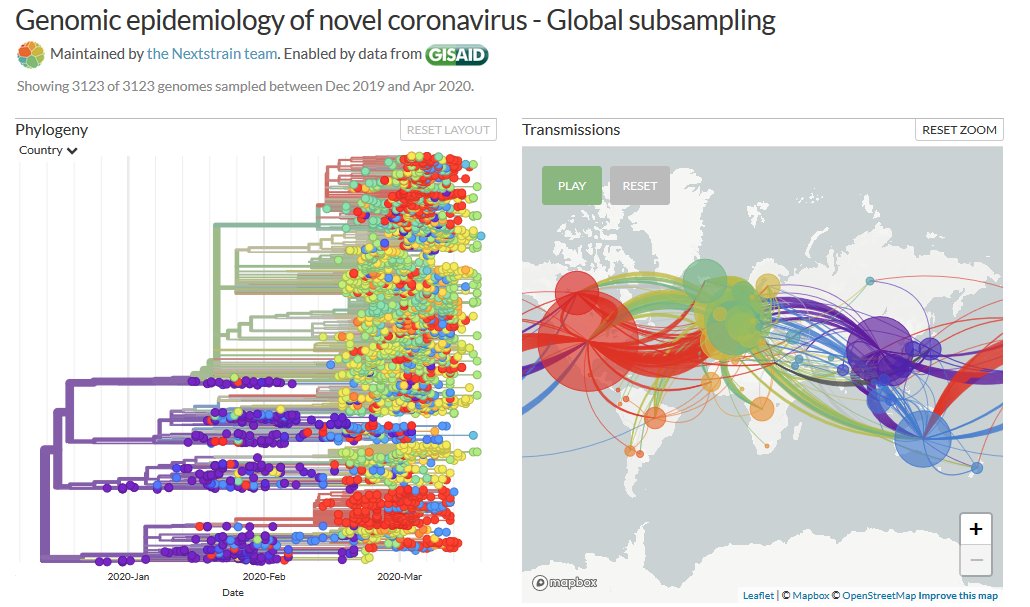
Thanks to #opendata sharing by @GISAID, we've updated nextstrain.org/ncov with 4060 new #COVID19 #SARSCoV2 sequences!
Check out the new sequences from Australia (2) on nextstrain.org/ncov/oceania.
(Thanks to @NSWHPathology)
1/6
Check out the new sequences from Australia (2) on nextstrain.org/ncov/oceania.
(Thanks to @NSWHPathology)
1/6

@GISAID @NSWHPathology You can find the new sequences from Morocco (10), Bangladesh (155), Indonesia (6) and Israel (182) on nextstrain.org/ncov/africa and nextstrain.org/ncov/asia.
(Thanks to @um5rabat, @SolutionDna, @gksavarHQ, NIHRD Indonesia, Israel Central Virology Lab)
2/6

(Thanks to @um5rabat, @SolutionDna, @gksavarHQ, NIHRD Indonesia, Israel Central Virology Lab)
2/6


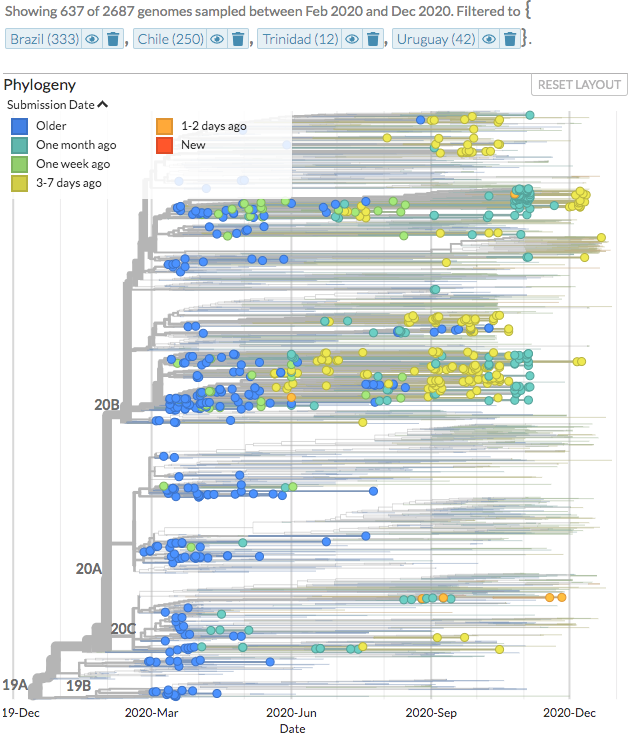
@GISAID @NSWHPathology @um5rabat @SolutionDna @gksavarHQ Check out the new sequences from Argentina (1) and Brazil (34) on nextstrain.org/ncov/south-ame….
(Thanks to @SADI_ORG, @lnccientifica, Instituto Adolfo Lutz)
3/6
(Thanks to @SADI_ORG, @lnccientifica, Instituto Adolfo Lutz)
3/6

@GISAID @NSWHPathology @um5rabat @SolutionDna @gksavarHQ @SADI_ORG @lnccientifica You can see the new sequences from the USA (399) on nextstrain.org/ncov/north-ame….
(Thanks to @NathanGrubaugh, @JosephFauver, @YaleSPH, @CDCgov, @MiGScenter, @LSUHS @LSUHSEVTLab, @MDHealthDept, @OHdeptofhealth, @UWVirology, @UW, Wyoming Public Health Lab, @pavitrarc)
4/6
(Thanks to @NathanGrubaugh, @JosephFauver, @YaleSPH, @CDCgov, @MiGScenter, @LSUHS @LSUHSEVTLab, @MDHealthDept, @OHdeptofhealth, @UWVirology, @UW, Wyoming Public Health Lab, @pavitrarc)
4/6

@GISAID @NSWHPathology @um5rabat @SolutionDna @gksavarHQ @SADI_ORG @lnccientifica @NathanGrubaugh @JosephFauver @YaleSPH @CDCgov @MiGScenter @LSUHS @LSUHSEVTLab @MDHealthDept @OHdeptofhealth @UWVirology @UW @pavitrarc New sequences from Germany (962), Ireland (25), Italy (306), & Netherlands (10) on nextstrain.org/ncov/europe.
(Thanks to @HeinrichPette, @UniklinikDUS @HHU_de, @nvrlucdireland, @teagasc Grange, @istitutotumori, L. Sacco @LaStatale, @Tigem_Telethon, IZSM Italy, @rivm)
5/6
(Thanks to @HeinrichPette, @UniklinikDUS @HHU_de, @nvrlucdireland, @teagasc Grange, @istitutotumori, L. Sacco @LaStatale, @Tigem_Telethon, IZSM Italy, @rivm)
5/6

@GISAID @NSWHPathology @um5rabat @SolutionDna @gksavarHQ @SADI_ORG @lnccientifica @NathanGrubaugh @JosephFauver @YaleSPH @CDCgov @MiGScenter @LSUHS @LSUHSEVTLab @MDHealthDept @OHdeptofhealth @UWVirology @UW @pavitrarc @HeinrichPette @UniklinikDUS @HHU_de @nvrlucdireland @teagasc @istitutotumori @LaStatale @Tigem_Telethon @rivm New sequences from Norway (262), Slovenia (1), Spain (12), Sweden (63) & United Kingdom (1630) can be found on nextstrain.org/ncov/europe.
(Thanks to @Folkehelseinst, NLZOH Slovenia, Hospital General Universitario Gregorio Marañón, @INIA_es, @Folkhalsomynd, @CovidGenomicsUK)
6/6
(Thanks to @Folkehelseinst, NLZOH Slovenia, Hospital General Universitario Gregorio Marañón, @INIA_es, @Folkhalsomynd, @CovidGenomicsUK)
6/6

• • •
Missing some Tweet in this thread? You can try to
force a refresh