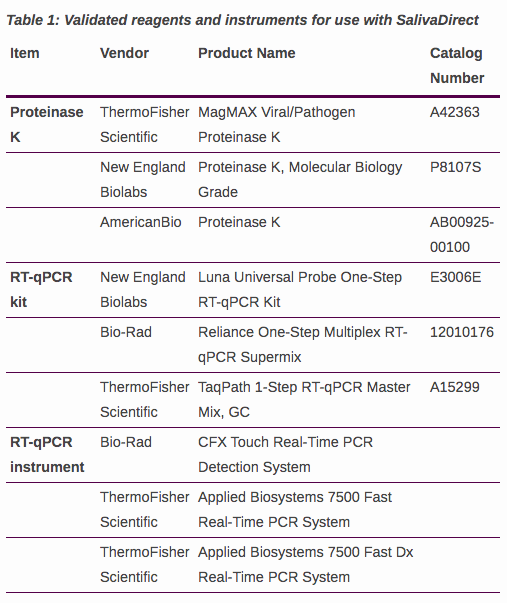
Brief update on TaqPath N gene target failure (NGTF). We believe that it is caused by a 6nt deletion (positions 28914-28919; N:214/215del) that has emerged independently at least 3 times in the US.
short 🧵
nextstrain.org/community/grub…
short 🧵
nextstrain.org/community/grub…
https://twitter.com/NathanGrubaugh/status/1349704743564832770
The @yalepathology clinical diagnostic lab recently detected 8 NGTFs from a related cluster of cases. We have so far sequenced 2 of these (Yale-S047 & Yale-S048) & they do not cluster with other N:214/215del seqs. Rather looks like it emerged within a local lineage (B.1.1.107). 

33 N:214/215del seqs are located in the Pango lineage B.1.3, and have been detected in New York and Maryland. 

23 N:214/215del seqs are located in a yet-to-be assigned B.1 Pango lineage (bat-signaling @AineToole) sequenced from Florida. 

We don't know of any phenotypic effects of N:214/215del, but given its impact on the TaqPath - @thermofisher (@ThermoFisherPR) and users of this assay should be aware.
• • •
Missing some Tweet in this thread? You can try to
force a refresh