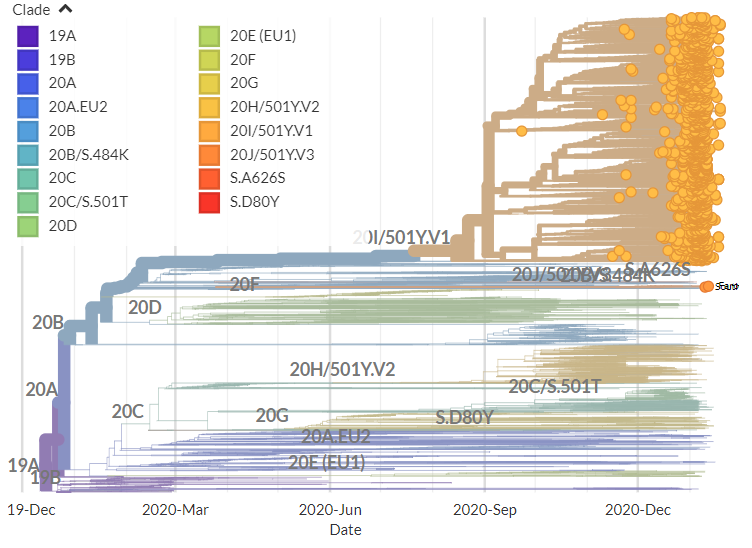
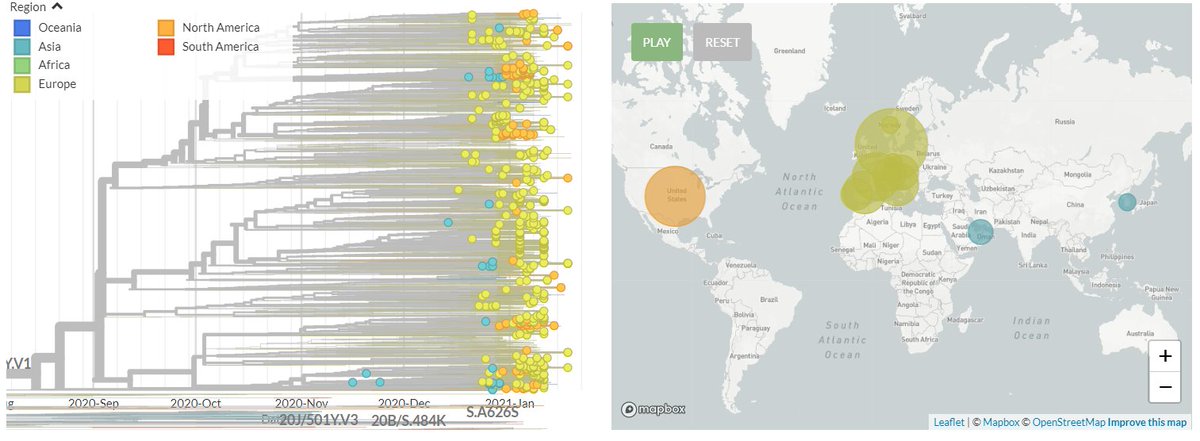
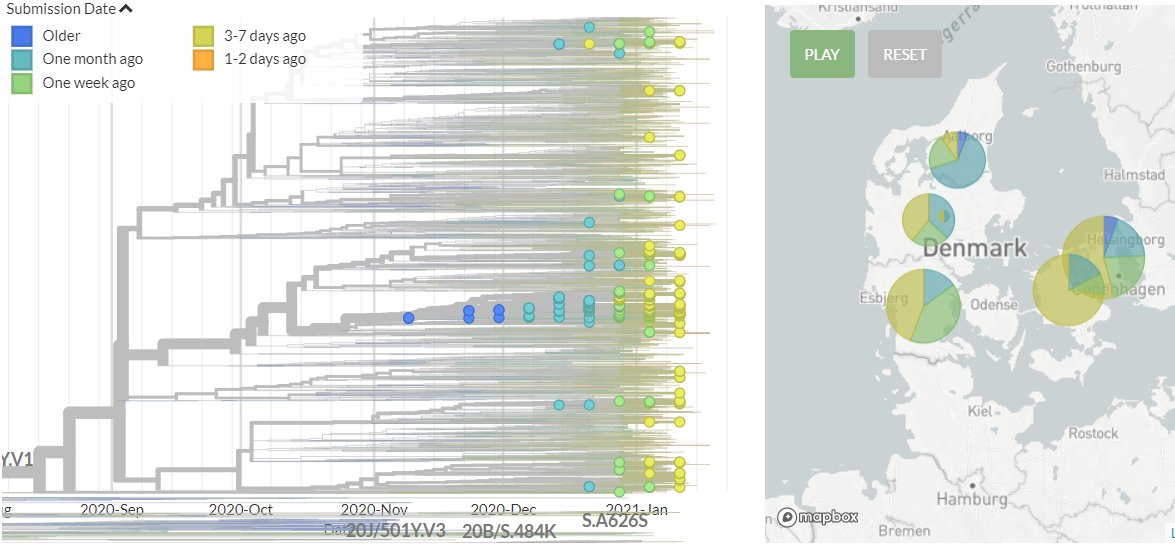
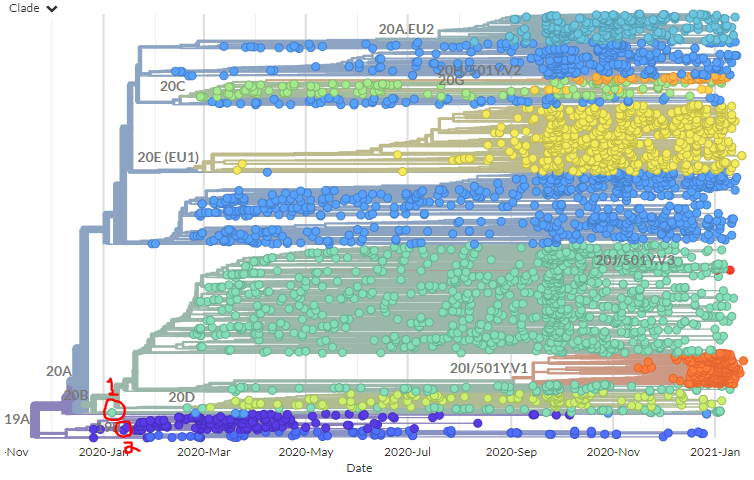
🎉The new and improved CoVariants.org is here! 🎉
We're hoping to make this a one-stop location to track not only the 'headline' variants, but many different #SARSCoV2 mutations & variants that scientists are keeping an eye on.
Let's take a tour of the site!
1/12
We're hoping to make this a one-stop location to track not only the 'headline' variants, but many different #SARSCoV2 mutations & variants that scientists are keeping an eye on.
Let's take a tour of the site!
1/12
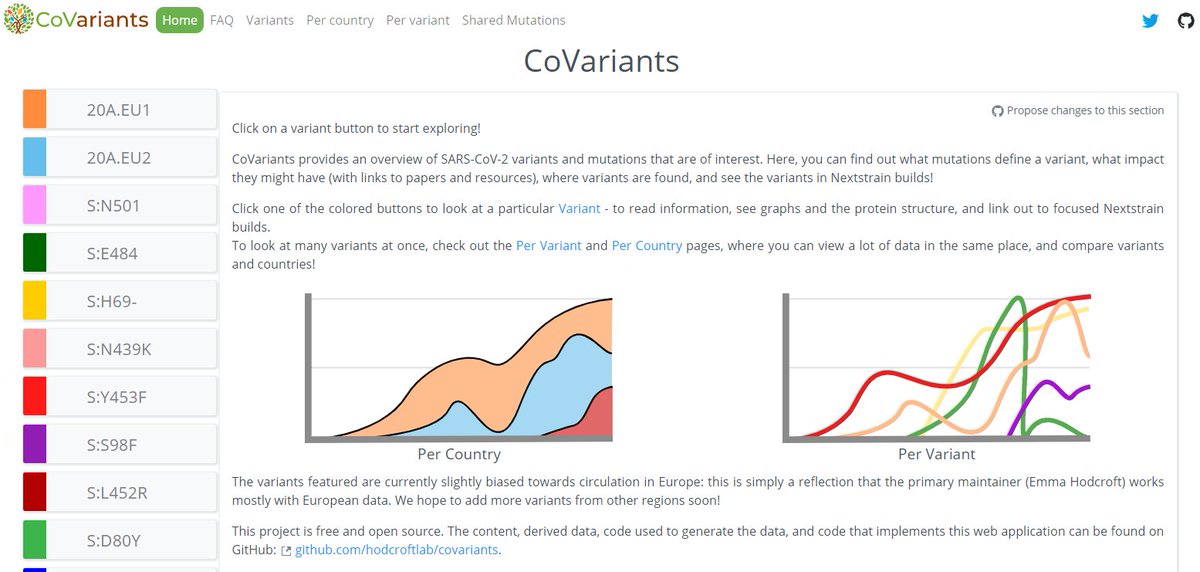
The buttons on the left will take you to pages focused on key variants & mutations, while the menu at the top will help you jump between info pages (FAQ) & pages compiling together information about many variants (ex: Per country).
Let's take a look at the 20A.EU1 page
2/12
Let's take a look at the 20A.EU1 page
2/12
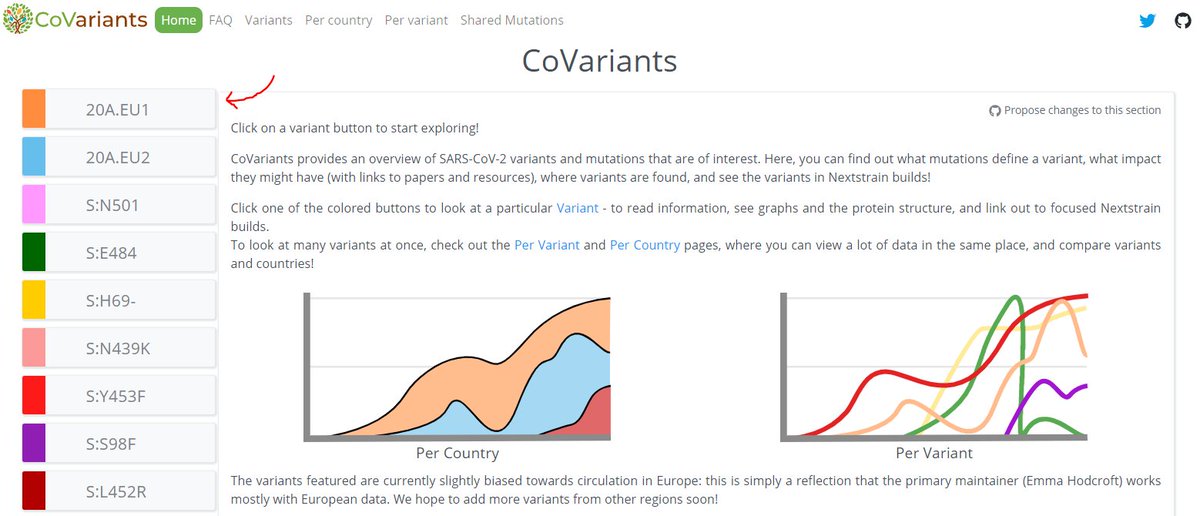
Here, we can
- Click through to a dedicated @Nextstrain build for the variant (1)
- See a list of the defining mutations (2)
- Learn a bit about the variant & see links to papers (3)
- See a distribution of 20A.EU1 per country (4)
- See a protein model! (pic 2)
3/12

- Click through to a dedicated @Nextstrain build for the variant (1)
- See a list of the defining mutations (2)
- Learn a bit about the variant & see links to papers (3)
- See a distribution of 20A.EU1 per country (4)
- See a protein model! (pic 2)
3/12

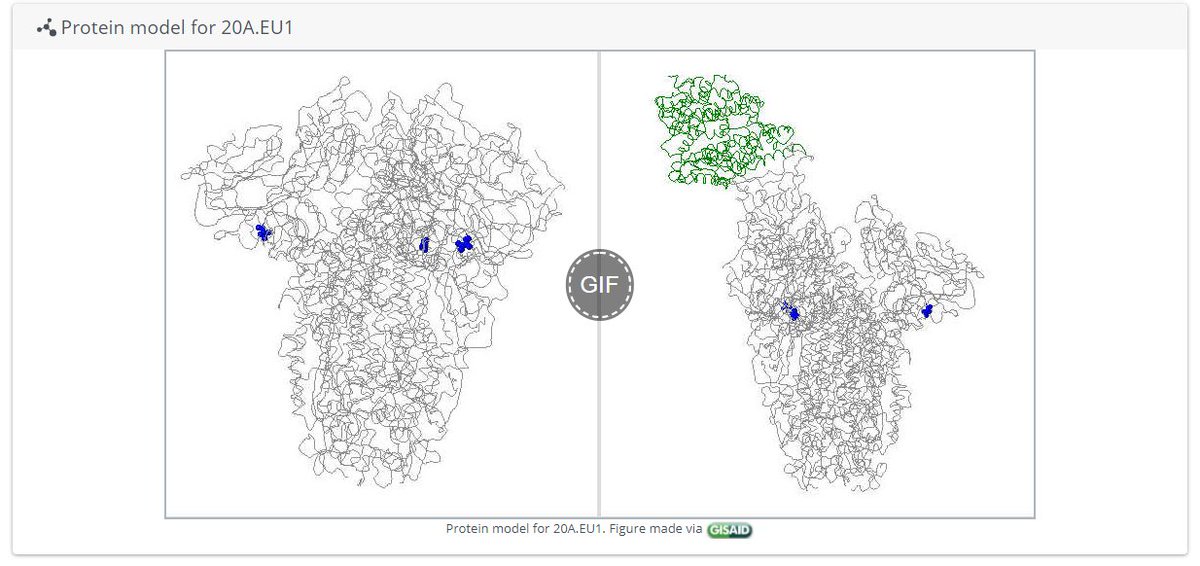
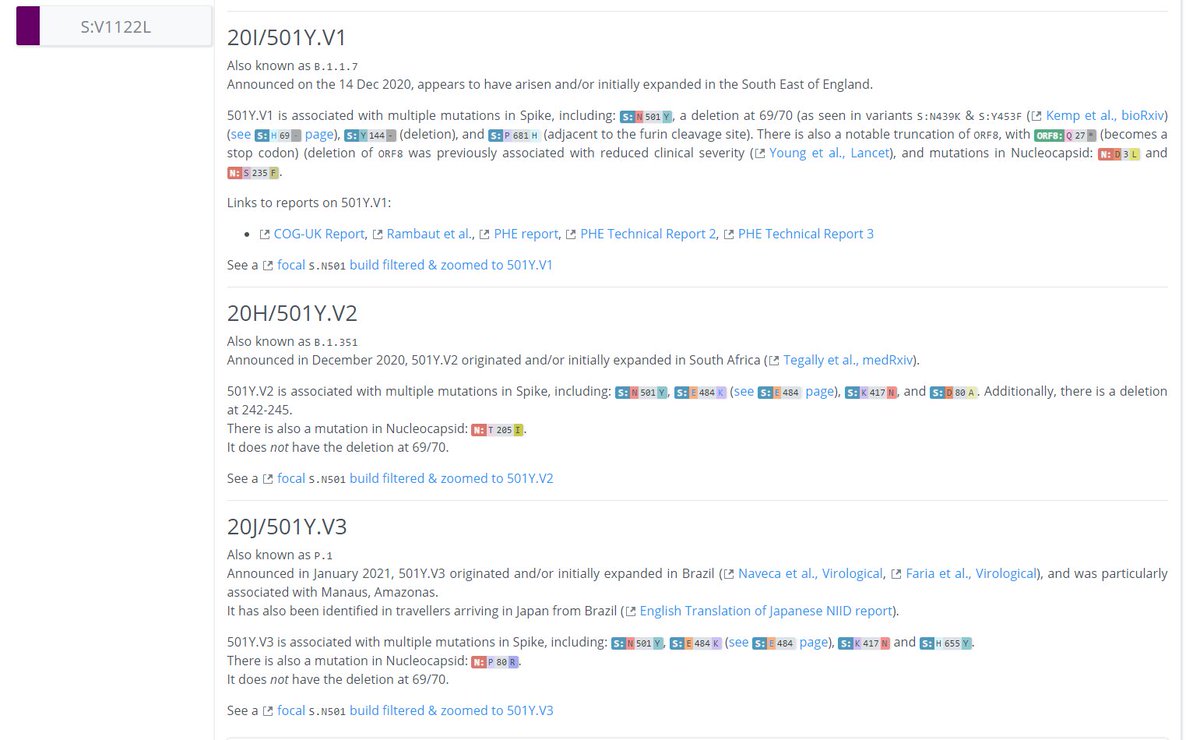
On other pages we can see even more information.
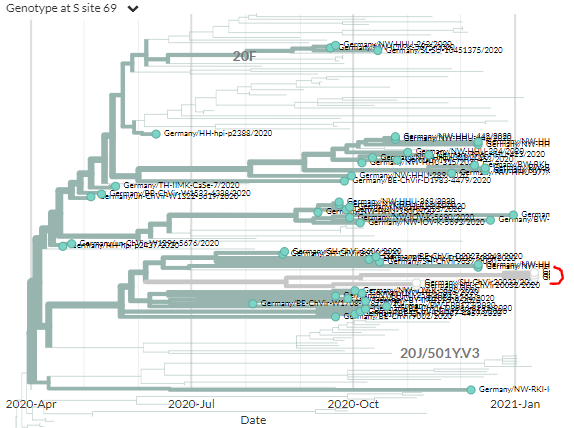
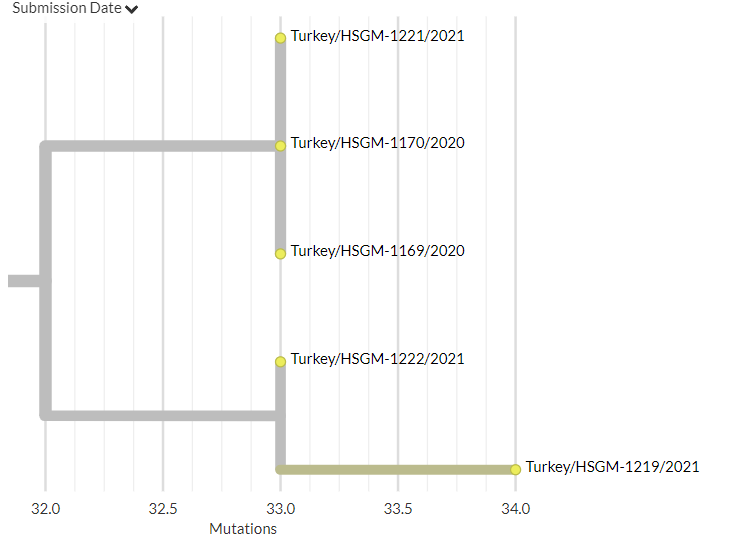
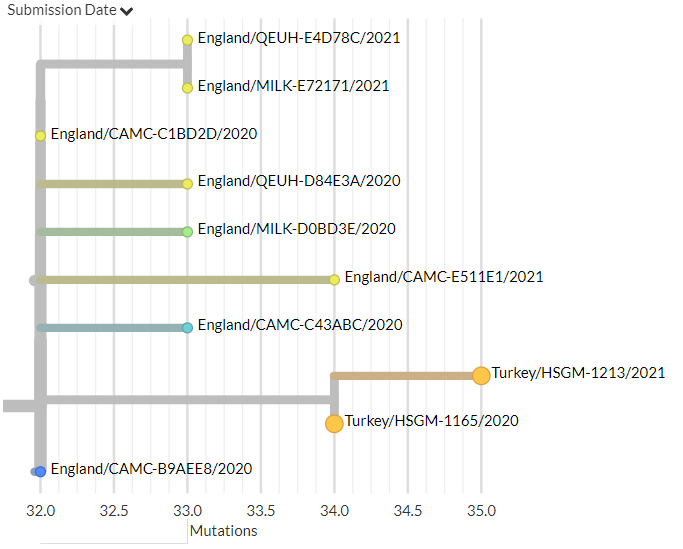
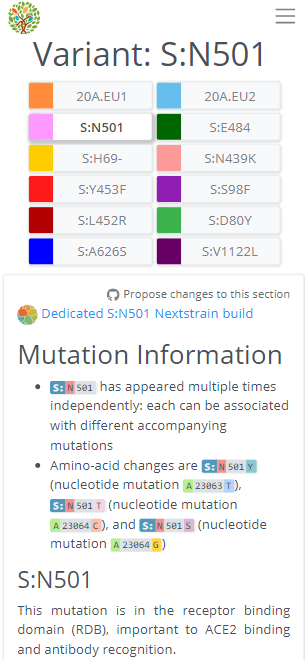
On the S:N501 page, we can see even more information & links about the S:N501 mutation itself (pic 1), & detailed information on the 3 variants that have this mutation (501Y.V1 [B.1.1.7], 501Y.V2, 501Y.V3) (pic 2).
4/12

On the S:N501 page, we can see even more information & links about the S:N501 mutation itself (pic 1), & detailed information on the 3 variants that have this mutation (501Y.V1 [B.1.1.7], 501Y.V2, 501Y.V3) (pic 2).
4/12

Often we're curious about the trends of these variants in particular countries. We can check out 'Per country' for an interactive overview of the variant frequencies in select countries.
You can mouse-over to find out the sequence counts, & turn on/off variants!
5/12
You can mouse-over to find out the sequence counts, & turn on/off variants!
5/12
We can also look at information 'Per Variant' with what frequency of each country's sequences fall in that variant.
Again, mousing over give more detailed information, and countries can be turned on and off!
6/12
Again, mousing over give more detailed information, and countries can be turned on and off!
6/12
Finally - we've added a table where you can easily see what mutations are shared between some of the variants getting the most important attention right now. Use the toggle to change how the mutations are sorted!
We'll expand this table in future!
7/12
We'll expand this table in future!
7/12
You can find out more information about the this website - FAQs, citation & data information, useful resources, and more - in the FAQ pages!
8/12
8/12

The #SARSCoV2 pandemic & the research on it is ongoing & changes every day. We're making every effort to keep CoVariants updated, but welcome PR requests & issues to ensure it is as useful as possible!
See links at the top right of each section!
9/12
github.com/hodcroftlab/co…
See links at the top right of each section!
9/12
github.com/hodcroftlab/co…
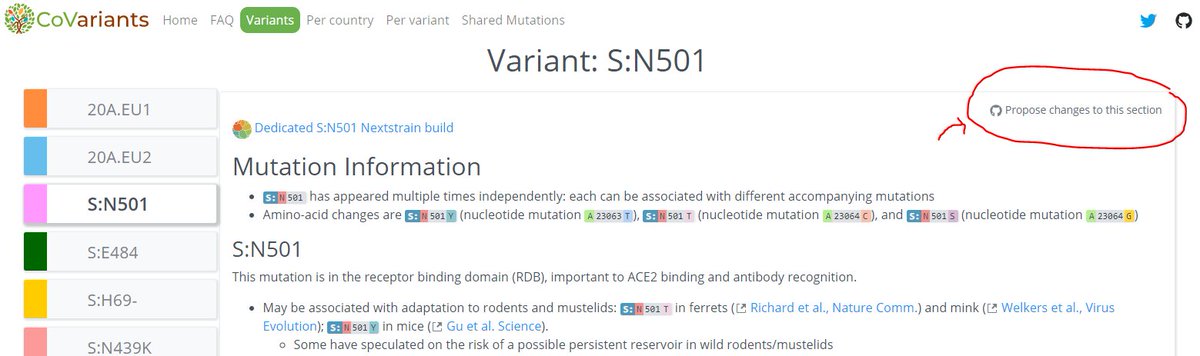
Further, we've already got a *lot* of updates in the works, including a better breakdown of the variants (buttons for each one!) & more variants in the mutation table!
We hope to make CoVariants even more useful for #SARSCoV2 communication & research.
10/12
We hope to make CoVariants even more useful for #SARSCoV2 communication & research.
10/12
CoVariants is also mobile & tablet friendly 📱, so you can be sure to keep up with the latest graphs & information even on-the-go! 😆😉
11/12

11/12

And of course, this work was not done alone. Huge thanks to everyone who's helped so far.
In particular, massive 👏🏻👏🏻 to @ivan_aksamentov for the web design.
Also big thanks to @hamesjadfield for the mutation table idea, & @richardneher & @trvrb for feedback & support!
12/12
In particular, massive 👏🏻👏🏻 to @ivan_aksamentov for the web design.
Also big thanks to @hamesjadfield for the mutation table idea, & @richardneher & @trvrb for feedback & support!
12/12
PS - Don't forget to update your bookmarks! CoVariants.org will take you straight to the new page, but the Github (github.com/hodcroftlab/co…) has changed, so you'll have to click through.
To go straight there, point your bookmark to CoVariants.org!
To go straight there, point your bookmark to CoVariants.org!

• • •
Missing some Tweet in this thread? You can try to
force a refresh