As we head deeper into 2021, a gentle reminder to double-check your #SARSCoV2 sequencing scripts for hardcoded '2020's! 😬
We've seen a real uptick recently in sequences with sampling dates in Jan 2020, with divergence that clearly indicates they meant 2021! 📅🤦🏻♀️
We've seen a real uptick recently in sequences with sampling dates in Jan 2020, with divergence that clearly indicates they meant 2021! 📅🤦🏻♀️
"How can we tell?" you might wonder. Actually, these cases are pretty easy to spot!
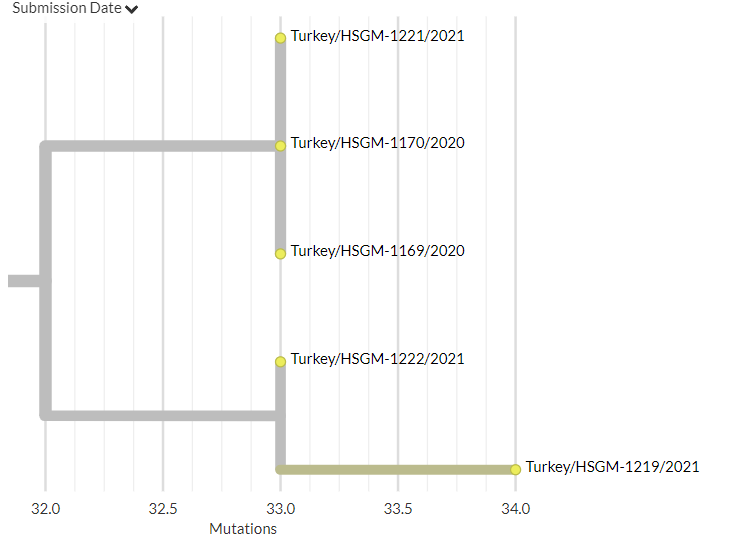
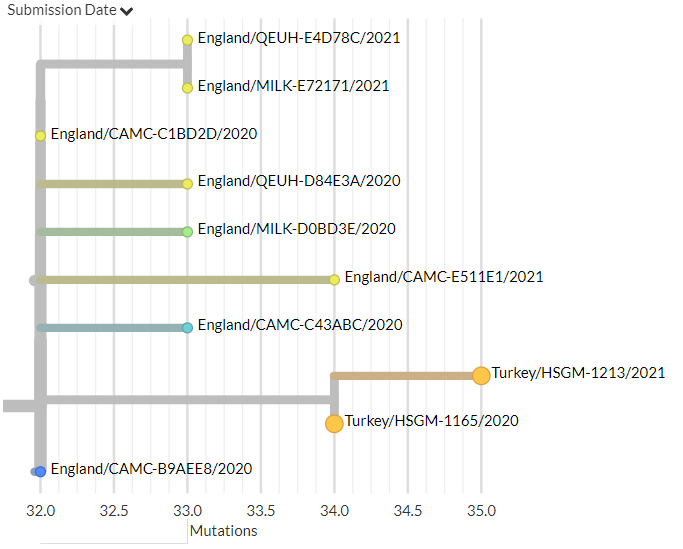
Let's take a look at two sequences reported as being from Jan 2020 - 1 in green, 2 in blue. (Spoiler, green is the imposter.)

Let's take a look at two sequences reported as being from Jan 2020 - 1 in green, 2 in blue. (Spoiler, green is the imposter.)


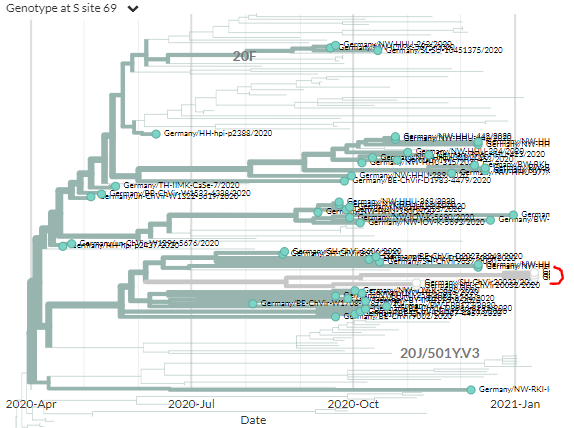
This view is 'date view' where sequences are plotted on the X-axis by the date they were sampled. Then, we try and infer how they're connected using their genetic information - specifically, what mutations they share & don't share. 
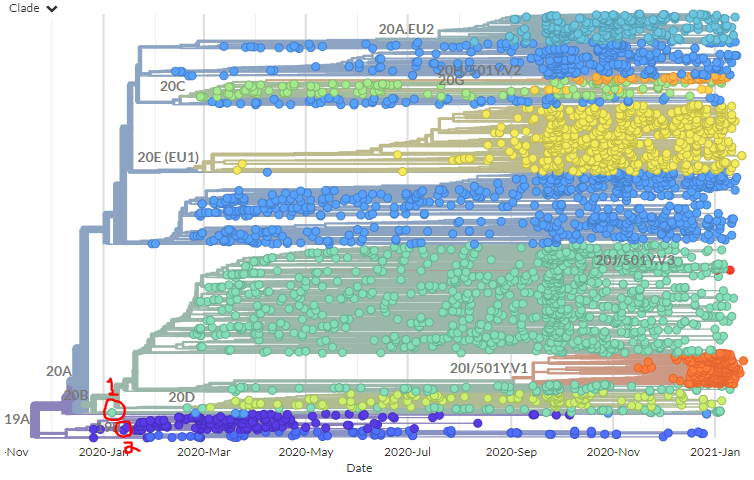
But we can Also view the tree in 'divergence view' - *just* looking at the mutation differences and not considering time.
Colouring by sample date, you can see that generally, recent sequences (red) have higher divergence (further to right).
Colouring by sample date, you can see that generally, recent sequences (red) have higher divergence (further to right).

In this view, the blue #2 sequence is wayyy to the left - as we'd expect from a sequence from Jan 2020.
But our friend green #1 - supposedly from the same time - is sitting in the middle, 20 muts away from #2!
(This fits a lot closer to the ~24 muts we'd expect for Jan 2021!)

But our friend green #1 - supposedly from the same time - is sitting in the middle, 20 muts away from #2!
(This fits a lot closer to the ~24 muts we'd expect for Jan 2021!)


We can also look at this in 'clock view' - plotting sample date (x-axis) against # of mutations (y-axis). We expect - on average - for sequences to lie roughly around the black line. 

But you can see imposter green sitting wayy too high, far from the line.
Given it's at 20 mutations, if we trace that y-axis line to the right, it crosses the black line at around the end of 2020. (However, mutation rates are an average - there's noise, so Jan 2021 also fits.)
Given it's at 20 mutations, if we trace that y-axis line to the right, it crosses the black line at around the end of 2020. (However, mutation rates are an average - there's noise, so Jan 2021 also fits.)

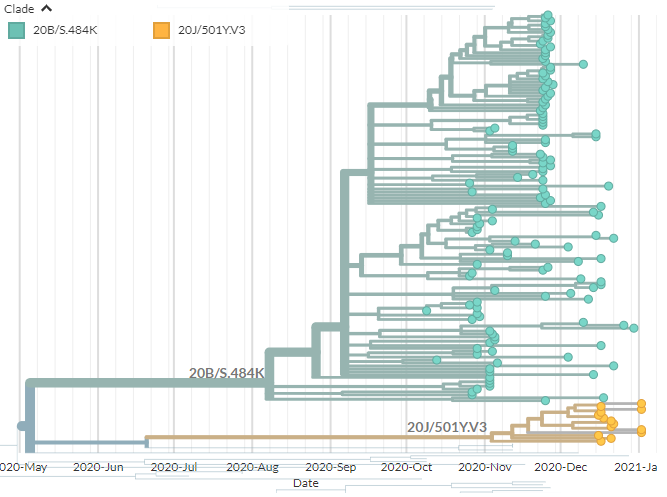
These views can also illustrate what you may have heard about 501Y.V1 (B.1.1.7) & 501Y.V3 having 'long branches' or 'many mutations.
Look at them in divergence & clock view. Even though the samples are mostly from end-2020/Jan-2021, they have 30+ mutations.

Look at them in divergence & clock view. Even though the samples are mostly from end-2020/Jan-2021, they have 30+ mutations.


How these long branches happen and what it means for #SARSCoV2 is greatly discussed by scientists, but I hope now if you hear it again, you can picture how scientists 'measure' this and why it 'stands out' so much!
And, I hope this mini-thread as helped you learn a little about how we can sometimes spot bad dates (and bad sequences!) in @Nextstrain runs - and a few things about how our analyses work, and some interesting ways you can view things on Nextstrain!
• • •
Missing some Tweet in this thread? You can try to
force a refresh