@TheSeeker268 @Daoyu15 @Undergroundsar3 @Real_Adam_B @BillyBostickson @Rossana38510044 @luigi_warren @babarlelephant @AntGDuarte @MonaRahalkar @flavinkins @KevinMcH3 @DrAntoniSerraT1 @_coltseavers @rowanjacobsen @uacjess @RolandBakerIII @still_a_nerd @jjcouey @Harvard2H @ydeigin @CarltheChippy @ico_dna @nerdhaspower @scottburke777 @JJ2000426 @BahulikarRahul @alimhaider @antonioregalado @Ayjchan @R_H_Ebright @BretWeinstein @sanchak74 @JCalvertST @PeterDaszak @TheSeeker @nature @threadreaderapp
@TheSeeker268 @Daoyu15 @Undergroundsar3 @Real_Adam_B @BillyBostickson @Rossana38510044 @luigi_warren @babarlelephant @AntGDuarte @MonaRahalkar @flavinkins @KevinMcH3 @DrAntoniSerraT1 @_coltseavers @rowanjacobsen @uacjess @RolandBakerIII @still_a_nerd @jjcouey @Harvard2H @ydeigin @CarltheChippy @ico_dna @nerdhaspower @scottburke777 @JJ2000426 @BahulikarRahul @alimhaider @antonioregalado @Ayjchan @R_H_Ebright @BretWeinstein @sanchak74 @JCalvertST @PeterDaszak @TheSeeker @nature @threadreaderapp This email seems to be an automatic email upon submission. It is dated as Jan 8th consistent with what the Genbank ID method suggests (Note: allow 1 day of difference due to timezones or slow batch processes). So, what is the other date of Jan 5th?
https://twitter.com/AntGDuarte/status/1358757270629269505
@TheSeeker268 @Daoyu15 @Undergroundsar3 @Real_Adam_B @BillyBostickson @Rossana38510044 @luigi_warren @babarlelephant @AntGDuarte @MonaRahalkar @flavinkins @KevinMcH3 @DrAntoniSerraT1 @_coltseavers @rowanjacobsen @uacjess @RolandBakerIII @still_a_nerd @jjcouey @Harvard2H @ydeigin @CarltheChippy @ico_dna @nerdhaspower @scottburke777 @JJ2000426 @BahulikarRahul @antonioregalado @Ayjchan @R_H_Ebright @BretWeinstein @sanchak74 @JCalvertST @PeterDaszak @TheSeeker @nature @threadreaderapp Could be this:
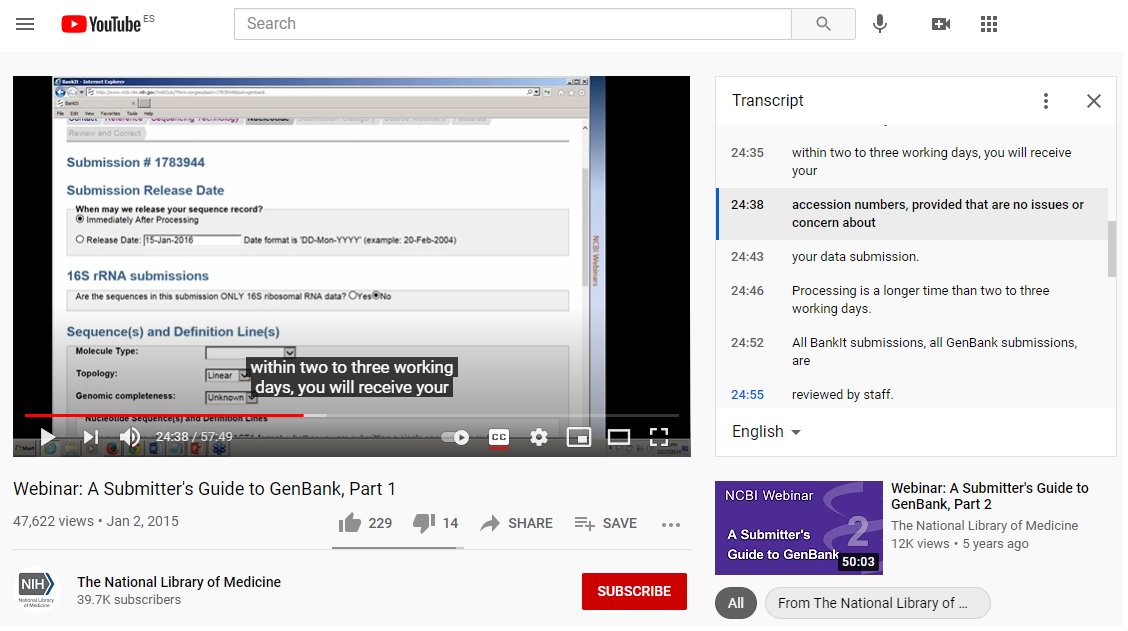
"GenBank will provide accession numbers for submitted sequences, usually within two working days"
NCBI still has many non automated processes, even receiving submissions by email. So that could explain those 3 days of difference
ncbi.nlm.nih.gov/genbank/submit/
"GenBank will provide accession numbers for submitted sequences, usually within two working days"
NCBI still has many non automated processes, even receiving submissions by email. So that could explain those 3 days of difference
ncbi.nlm.nih.gov/genbank/submit/

@TheSeeker268 @Daoyu15 @Undergroundsar3 @Real_Adam_B @BillyBostickson @Rossana38510044 @luigi_warren @babarlelephant @AntGDuarte @MonaRahalkar @flavinkins @KevinMcH3 @DrAntoniSerraT1 @_coltseavers @rowanjacobsen @uacjess @RolandBakerIII @still_a_nerd @jjcouey @Harvard2H @ydeigin @CarltheChippy @ico_dna @nerdhaspower @scottburke777 @JJ2000426 @BahulikarRahul @antonioregalado @Ayjchan @R_H_Ebright @BretWeinstein @sanchak74 @JCalvertST @PeterDaszak @TheSeeker @nature @threadreaderapp In ddition, it seems that the updating process could be even more manual. And in the cases of not being processed or released yet, I am not sure if we would see public traces of the updates in the records
ncbi.nlm.nih.gov/genbank/update/
ncbi.nlm.nih.gov/genbank/update/
@TheSeeker268 @Daoyu15 @Undergroundsar3 @Real_Adam_B @BillyBostickson @Rossana38510044 @luigi_warren @babarlelephant @AntGDuarte @MonaRahalkar @flavinkins @KevinMcH3 @DrAntoniSerraT1 @_coltseavers @rowanjacobsen @uacjess @RolandBakerIII @still_a_nerd @jjcouey @Harvard2H @ydeigin @CarltheChippy @ico_dna @nerdhaspower @scottburke777 @JJ2000426 @BahulikarRahul @antonioregalado @Ayjchan @R_H_Ebright @BretWeinstein @sanchak74 @JCalvertST @PeterDaszak @TheSeeker @nature @threadreaderapp The 9 virus of Li et al. (2020) were released on 31-Dec-19, but their Genbank ID was assigned on 27-Dec-19 (so submitted around 25-Dec-19). So, either: 1. they were under embargo 4 days (weird!), 2. they were under a longer embargo, but decided to release

https://twitter.com/franciscodeasis/status/1282365428044427270

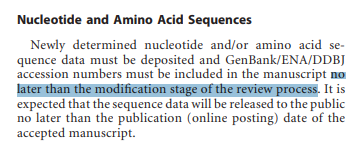
@TheSeeker268 @Daoyu15 @Undergroundsar3 @Real_Adam_B @BillyBostickson @Rossana38510044 @luigi_warren @babarlelephant @AntGDuarte @MonaRahalkar @flavinkins @KevinMcH3 @DrAntoniSerraT1 @_coltseavers @rowanjacobsen @uacjess @RolandBakerIII @still_a_nerd @jjcouey @Harvard2H @ydeigin @CarltheChippy @ico_dna @nerdhaspower @scottburke777 @JJ2000426 @BahulikarRahul @antonioregalado @Ayjchan @R_H_Ebright @BretWeinstein @sanchak74 @JCalvertST @PeterDaszak @TheSeeker @nature @threadreaderapp Although that paper was sent for peer review on 06-Nov-19 (finally published on 29-Jan-20), it seems that this journals seems to allow sending Accession "no later than the modification stage of the review process". It was accepted on 19-Dec-19
msphere.asm.org/sites/default/…
msphere.asm.org/sites/default/…
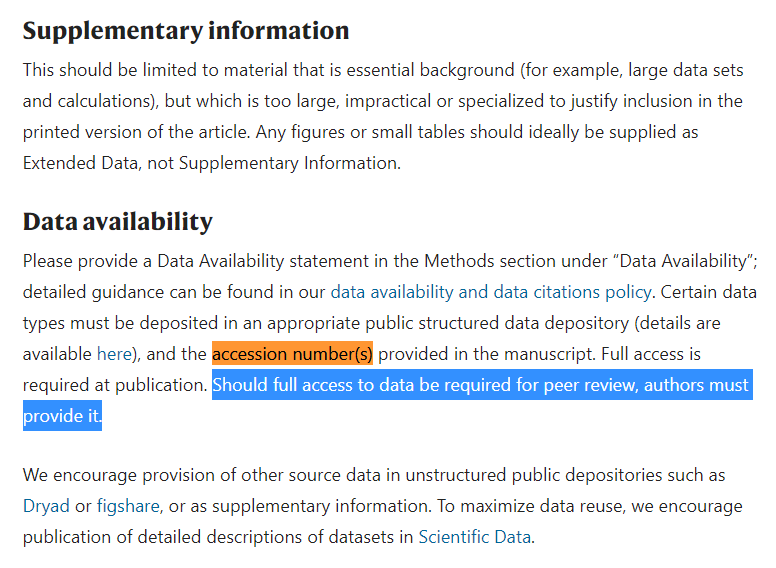
@TheSeeker268 @Daoyu15 @Undergroundsar3 @Real_Adam_B @BillyBostickson @Rossana38510044 @luigi_warren @babarlelephant @AntGDuarte @MonaRahalkar @flavinkins @KevinMcH3 @DrAntoniSerraT1 @_coltseavers @rowanjacobsen @uacjess @RolandBakerIII @still_a_nerd @jjcouey @Harvard2H @ydeigin @CarltheChippy @ico_dna @nerdhaspower @scottburke777 @JJ2000426 @BahulikarRahul @antonioregalado @Ayjchan @R_H_Ebright @BretWeinstein @sanchak74 @JCalvertST @PeterDaszak @TheSeeker @nature @threadreaderapp Regarding Latinne et al. (2020), published in Nature Com., the "Initial Submission" guide states that Accession numbers have to be provided, even with full access if necessary. So, what was sent on 06-Oct-19 one month before the accessions where created?
nature.com/nature/for-aut…
nature.com/nature/for-aut…
@TheSeeker268 @Daoyu15 @Undergroundsar3 @Real_Adam_B @BillyBostickson @Rossana38510044 @luigi_warren @babarlelephant @AntGDuarte @MonaRahalkar @flavinkins @KevinMcH3 @DrAntoniSerraT1 @_coltseavers @rowanjacobsen @uacjess @RolandBakerIII @still_a_nerd @jjcouey @Harvard2H @ydeigin @CarltheChippy @ico_dna @nerdhaspower @scottburke777 @JJ2000426 @BahulikarRahul @antonioregalado @Ayjchan @R_H_Ebright @BretWeinstein @sanchak74 @JCalvertST @PeterDaszak @TheSeeker @nature @threadreaderapp After discussing with @gdemaneuf, I see 3 possibilities: 1. Accessions manually updated by NCBI (& GI updated) at request without traces, 2. a first batch was withdrawn and they upload the current one (reusing Accessions), 3. they promised Nature to send the Accessions later
@TheSeeker268 @Daoyu15 @Undergroundsar3 @Real_Adam_B @BillyBostickson @Rossana38510044 @luigi_warren @babarlelephant @AntGDuarte @MonaRahalkar @flavinkins @KevinMcH3 @DrAntoniSerraT1 @_coltseavers @rowanjacobsen @uacjess @RolandBakerIII @still_a_nerd @jjcouey @Harvard2H @ydeigin @CarltheChippy @ico_dna @nerdhaspower @scottburke777 @JJ2000426 @BahulikarRahul @antonioregalado @Ayjchan @R_H_Ebright @BretWeinstein @sanchak74 @JCalvertST @PeterDaszak @TheSeeker @nature @threadreaderapp @gdemaneuf @gdemaneuf pointed the "submitted date" of "13-Aug-2019" as possible original submission date which is very plausible too.
The "submitted date" seems to be indicated by the submitter, but I think it is filled automatically if empty ncbi.nlm.nih.gov/nuccore/MN3126…
The "submitted date" seems to be indicated by the submitter, but I think it is filled automatically if empty ncbi.nlm.nih.gov/nuccore/MN3126…
https://twitter.com/franciscodeasis/status/1283536162171293696
@TheSeeker268 @Daoyu15 @Undergroundsar3 @Real_Adam_B @BillyBostickson @Rossana38510044 @luigi_warren @babarlelephant @AntGDuarte @MonaRahalkar @flavinkins @KevinMcH3 @DrAntoniSerraT1 @_coltseavers @rowanjacobsen @uacjess @RolandBakerIII @still_a_nerd @jjcouey @Harvard2H @ydeigin @CarltheChippy @ico_dna @nerdhaspower @scottburke777 @JJ2000426 @BahulikarRahul @antonioregalado @Ayjchan @R_H_Ebright @BretWeinstein @sanchak74 @JCalvertST @PeterDaszak @TheSeeker @nature @threadreaderapp @gdemaneuf Here the explanation: two ways of submitting data to Genbank:
- web-based tools: "submitted date" automatic.
- [local apps] preparation tools: "submitted date" (and any field) is filled by submitter
ncbi.nlm.nih.gov/genbank/submit/
And here how to do the embargo:


- web-based tools: "submitted date" automatic.
- [local apps] preparation tools: "submitted date" (and any field) is filled by submitter
ncbi.nlm.nih.gov/genbank/submit/
And here how to do the embargo:

@TheSeeker268 @Daoyu15 @Undergroundsar3 @Real_Adam_B @BillyBostickson @Rossana38510044 @luigi_warren @babarlelephant @AntGDuarte @MonaRahalkar @flavinkins @KevinMcH3 @DrAntoniSerraT1 @_coltseavers @rowanjacobsen @uacjess @RolandBakerIII @still_a_nerd @jjcouey @Harvard2H @ydeigin @CarltheChippy @ico_dna @nerdhaspower @scottburke777 @JJ2000426 @BahulikarRahul @antonioregalado @Ayjchan @R_H_Ebright @BretWeinstein @sanchak74 @JCalvertST @PeterDaszak @TheSeeker @nature @threadreaderapp @gdemaneuf So, it could be as @gdemaneuf suggested: something prepared on 13-Aug-2019 and either:
- was submitted then, sent to Nature and later withdrawn and replaced and re-sent to Nature
- was not submitted to Genbank until Nov 5th, so no Accession sent to Nature on time
- was submitted then, sent to Nature and later withdrawn and replaced and re-sent to Nature
- was not submitted to Genbank until Nov 5th, so no Accession sent to Nature on time
@TheSeeker268 @Daoyu15 @Undergroundsar3 @Real_Adam_B @BillyBostickson @Rossana38510044 @luigi_warren @babarlelephant @AntGDuarte @MonaRahalkar @flavinkins @KevinMcH3 @DrAntoniSerraT1 @_coltseavers @rowanjacobsen @uacjess @RolandBakerIII @still_a_nerd @jjcouey @Harvard2H @ydeigin @CarltheChippy @ico_dna @nerdhaspower @scottburke777 @JJ2000426 @BahulikarRahul @antonioregalado @Ayjchan @R_H_Ebright @BretWeinstein @sanchak74 @JCalvertST @PeterDaszak @TheSeeker @nature @threadreaderapp @gdemaneuf Regarding Wuhan-Hu-1 (MN908947).
Quod erat demonstrandum
Quod erat demonstrandum
https://twitter.com/franciscodeasis/status/1364269875796062216
• • •
Missing some Tweet in this thread? You can try to
force a refresh