
Endogenous tagging + 3D live-cell imaging + mass-spec interactome = opencell.czbiohub.org, an open-source map of the human proteome. So excited for our first release, wonderful collaboration with @labs_mann @loicaroyer & many other colleagues. @OpenCellCZB 
https://twitter.com/biorxivpreprint/status/1376693108512059396

(2/8) We used endogenous #CRISPR tagging with fluorescent probes to map both localization and interactions of proteins in their native cellular context. 

(3/8) We built a fully interactive website to share all of our data, including an intuitive 2D/3D viewer for microscopy images: opencell.czbiohub.org
Shout-out to @kchev for web design & engineering
#opendata #opensource
Shout-out to @kchev for web design & engineering
#opendata #opensource

(4/8) By encoding localization and interaction patterns for each protein, we applied unsupervised clustering methods to dissect the structure of the human proteome (similar to strategies used in single-cell RNASeq analysis) 
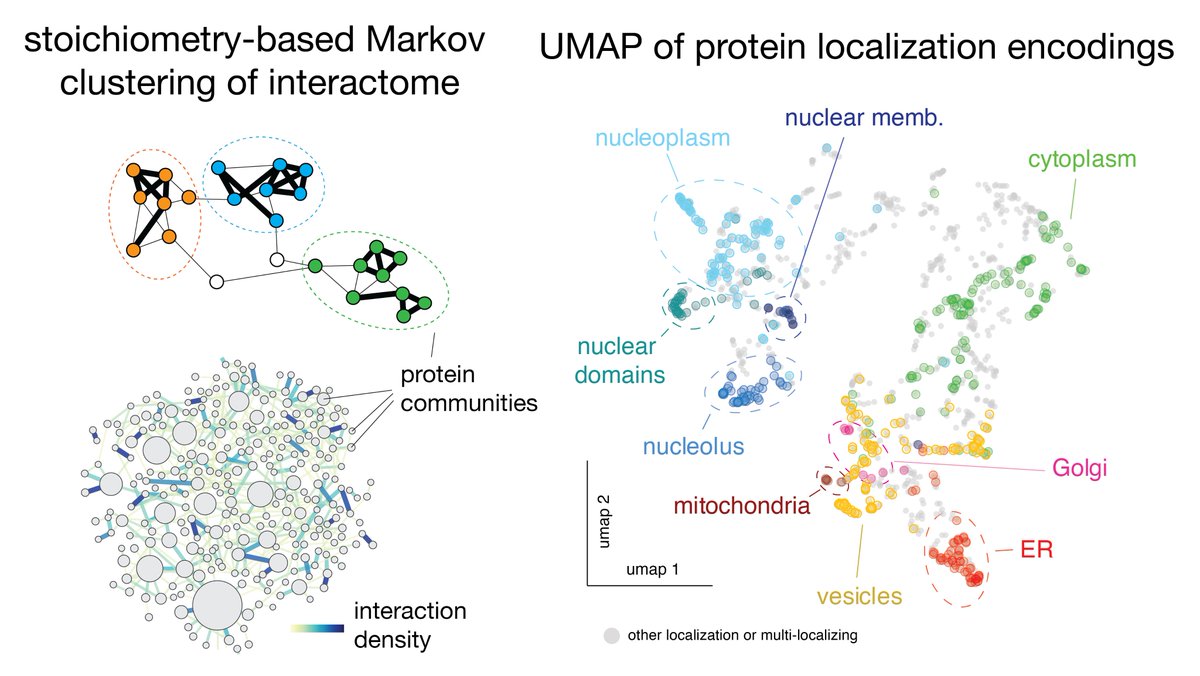
(5/8) Localization encodings are the first demonstration of #cytoself, our new self-supervised deep learning algorithm to encode protein localization from fluorescence imaging. Awesome work with @loicaroyer & @liilii_tweet. Check out our companion study
https://twitter.com/loicaroyer/status/1376702714474860548?s=20
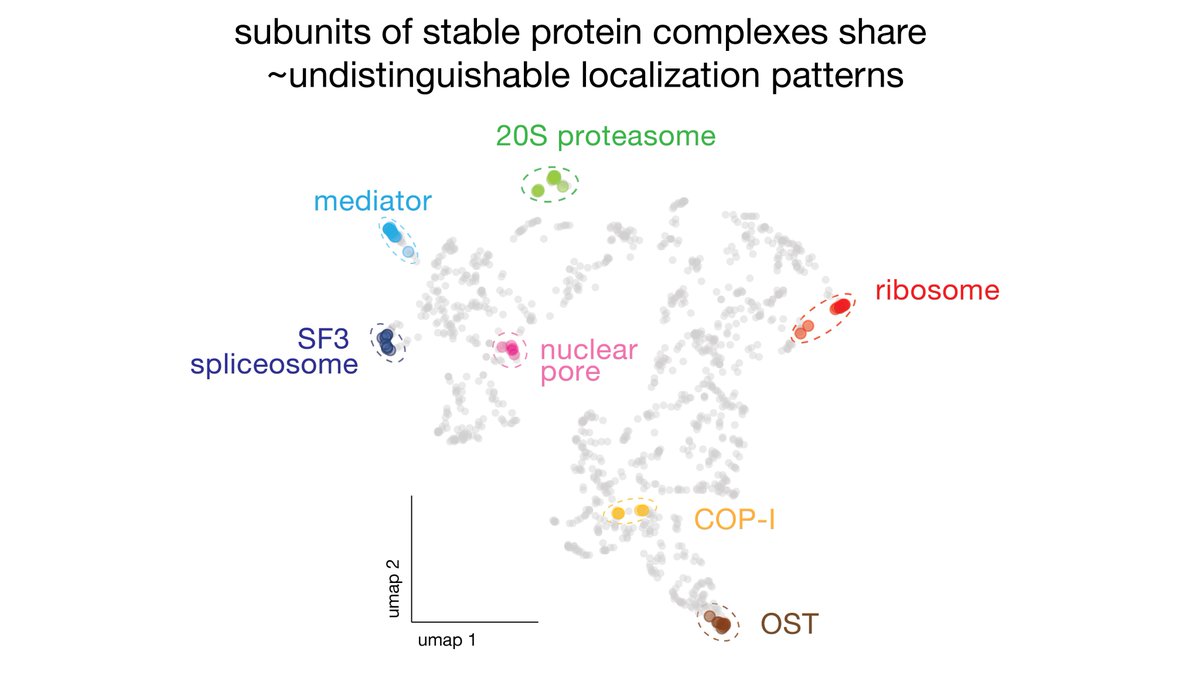
(6/8) We discover that localization signatures are remarkably predictive of protein function, and even contain enough information to identify molecular interactions. This is very interesting for the use of fluorescence microscopy for screening & functional genomics. 
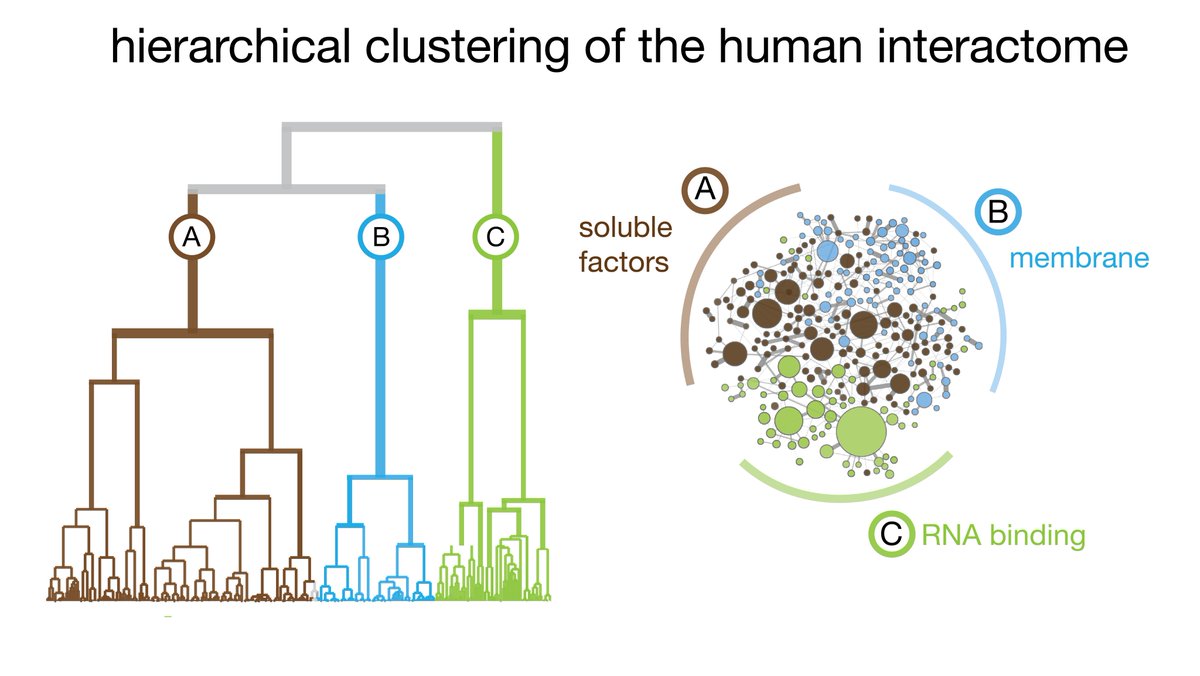
(7/8) All proteomics was done with @labs_mann - fantastic collaboration! Among many mechanistic insights, our interactome reveals that RNA-binding proteins segregate from the rest of the proteome (so do membrane-related proteins). Intrinsic protein disorder likely plays a role. 
(8/8) Check out our work and give us feedback! We hope that our resource will be useful for the community, and we’re always looking for ways to improve it.
• • •
Missing some Tweet in this thread? You can try to
force a refresh


