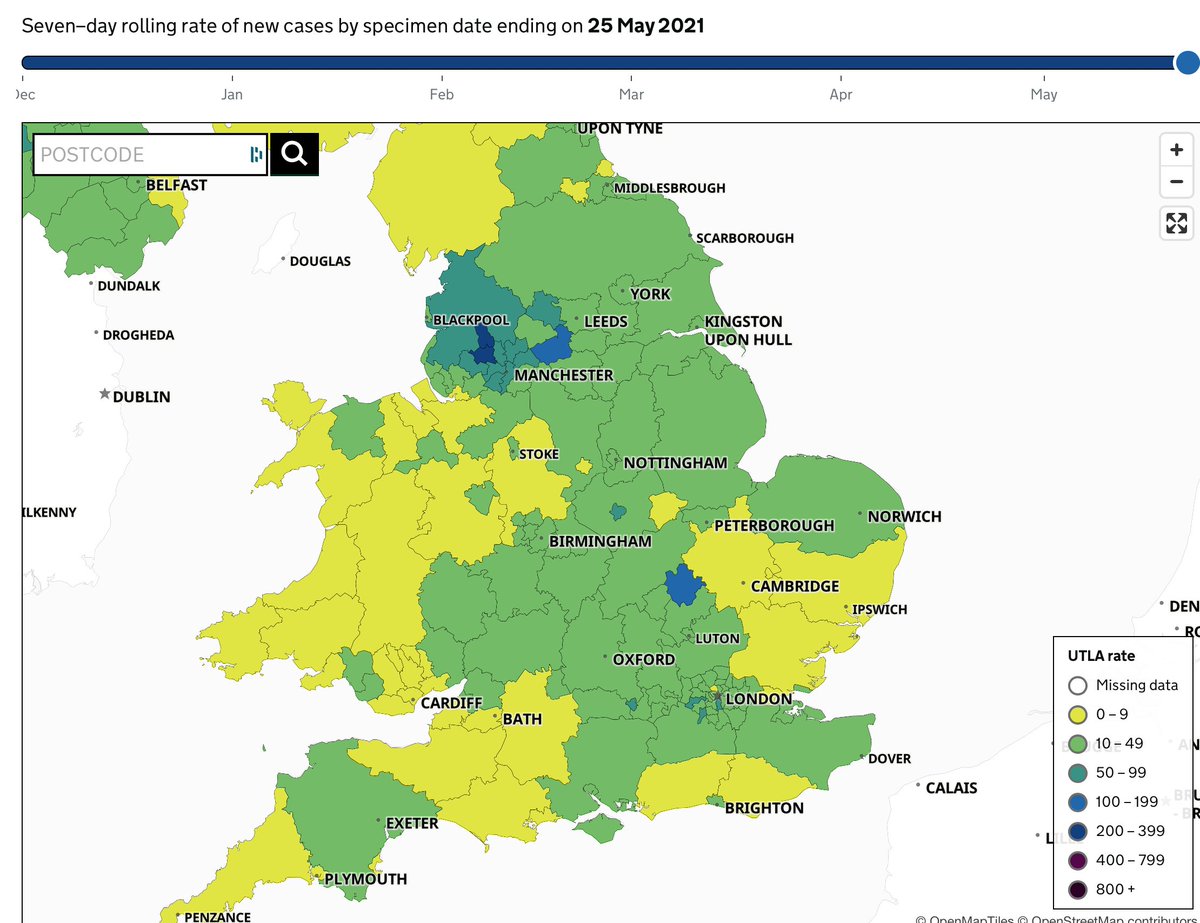
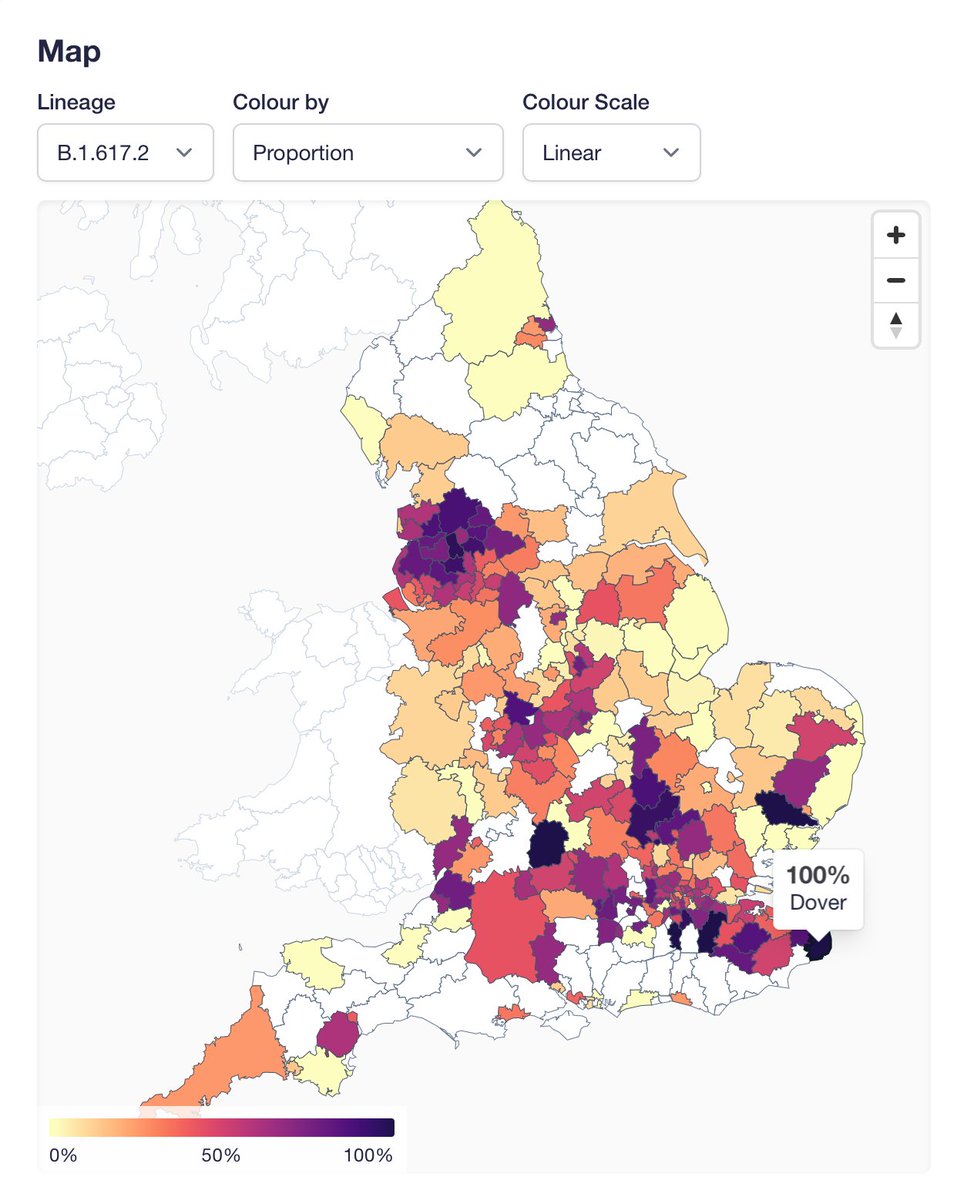
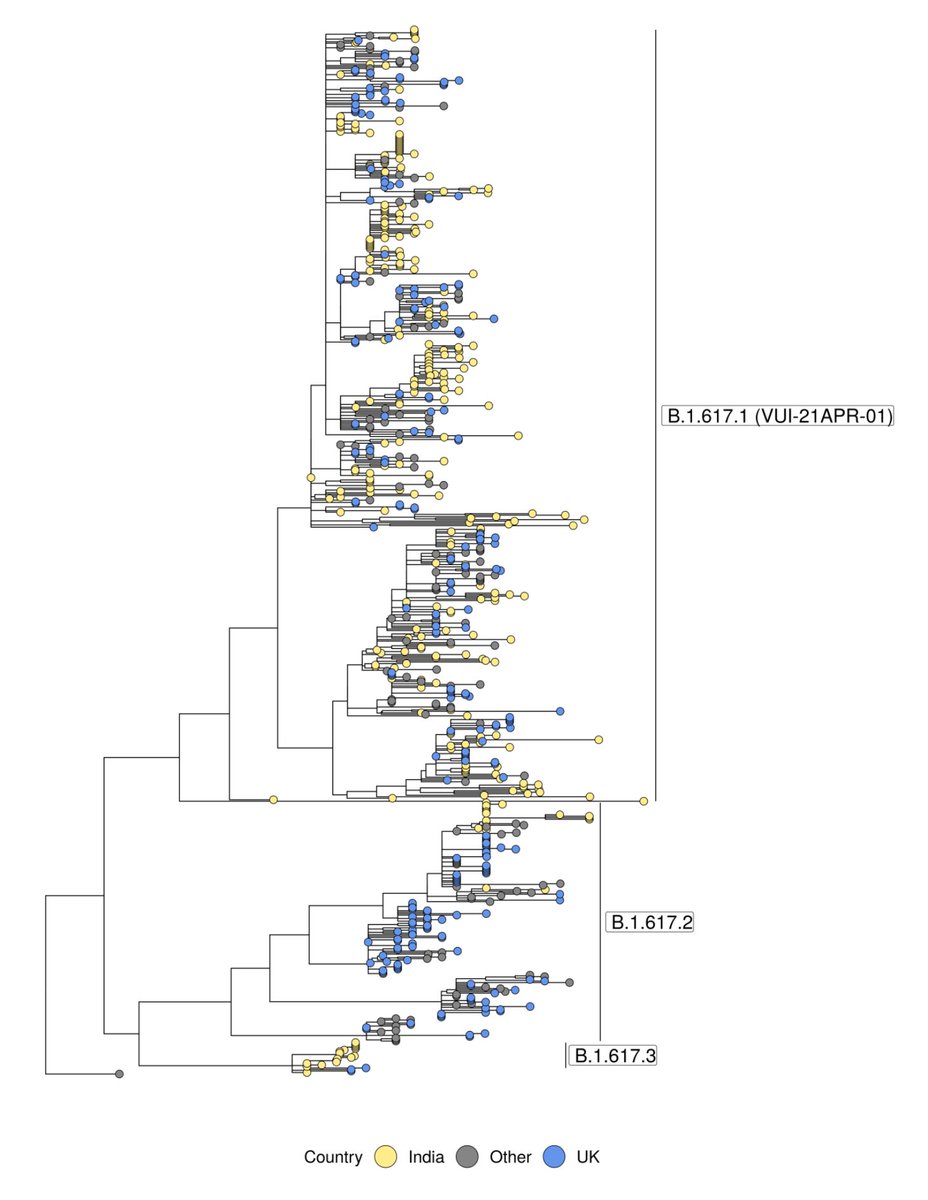
The description of genomic analyses in this piece about the spread of the Delta variant in the UK has major inaccuracies, and hindsight bias. 🧵
theguardian.com/commentisfree/…
theguardian.com/commentisfree/…
Early on, the author, @chrischirp, notes that Pakistan and Bangladesh were red-listed on 9 April, but India wasn't until 23 April. There is a reasonable criticism to be made here, but it has nothing to do with variants. Cases and % positivity were already on the rise in India 

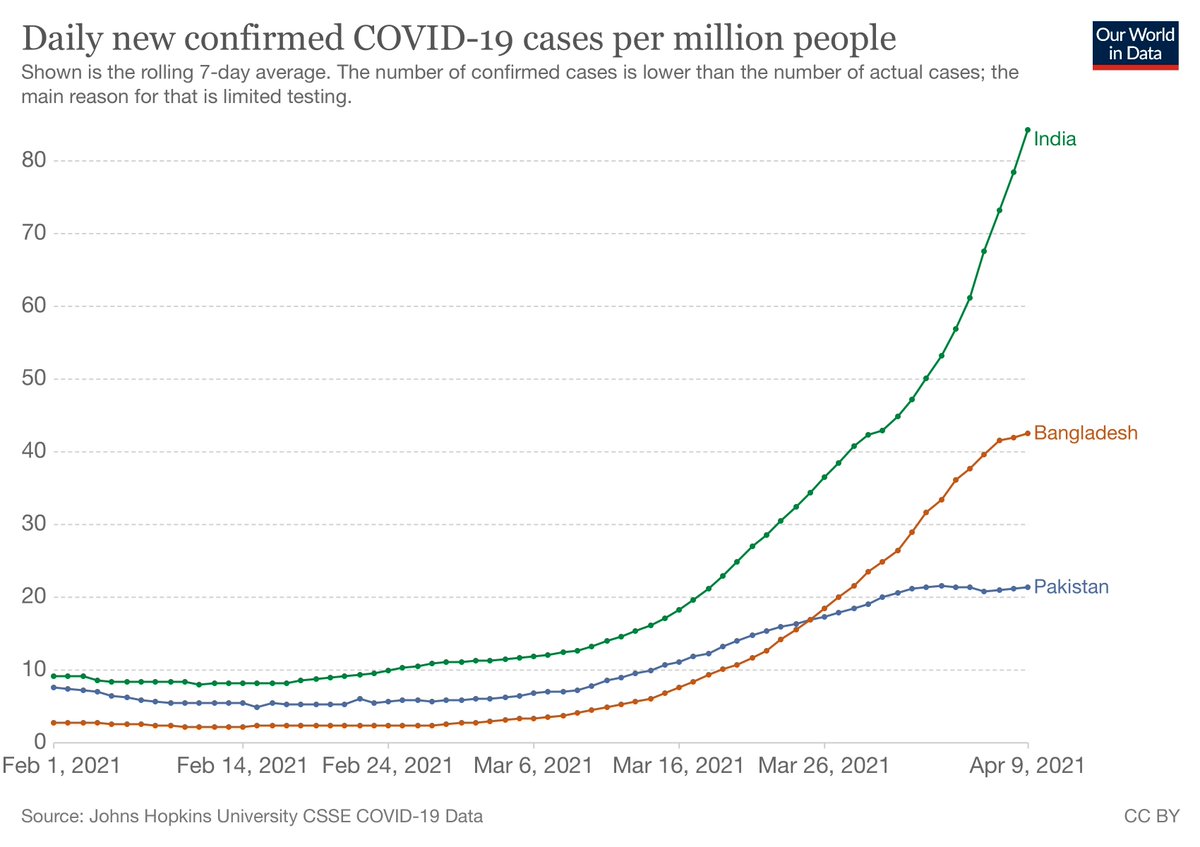

We know now that delay led to hundreds of importations of the Delta variant. The article says India "first reported a concerning new variant on 24 March...so why didn’t PHE immediately escalate Delta to a variant of concern in March?"
Well, first of all, the variant reported in late March was *not* Delta / B.1.617.2, but its cousin, B.1.617.1. At the time called (ridiculously) the "double mutant": L452R and E484Q. E484Q got a lot of attention about vaccine escape b/c we had all been watching E484K variants.
But in contrast to the statement in the article that "The new Delta variant has mutations that are associated with vaccine resistance", it was B.1.617.1 that got all the early attention! And IMHO, we still don't know which mutations give Delta its advantage. 
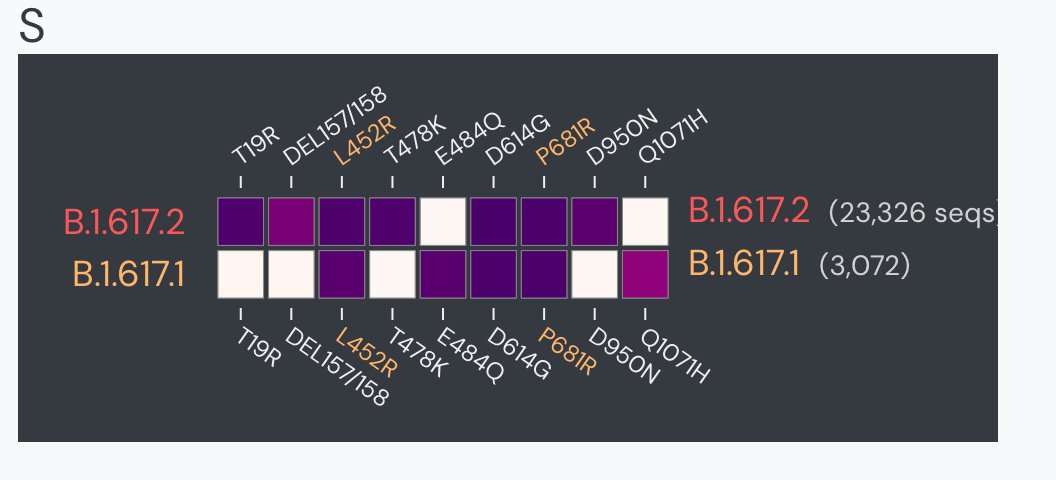
In fact, the three sub-lineages of B.1.617 were not even considered separately until 21 April, which is conveniently documented because the amazing Pango team (in this case @AineToole ) track lineage assignments in public github issues: github.com/cov-lineages/p…
So rather than being "obvious to anyone watching the relentless exponential growth of Delta during April.", it was actually pretty confusing. I make weekly VOC/VUI monitoring plots, and here's what I saw in three weeks from 19 April (1st time I included B.1.617) to 3 May. 



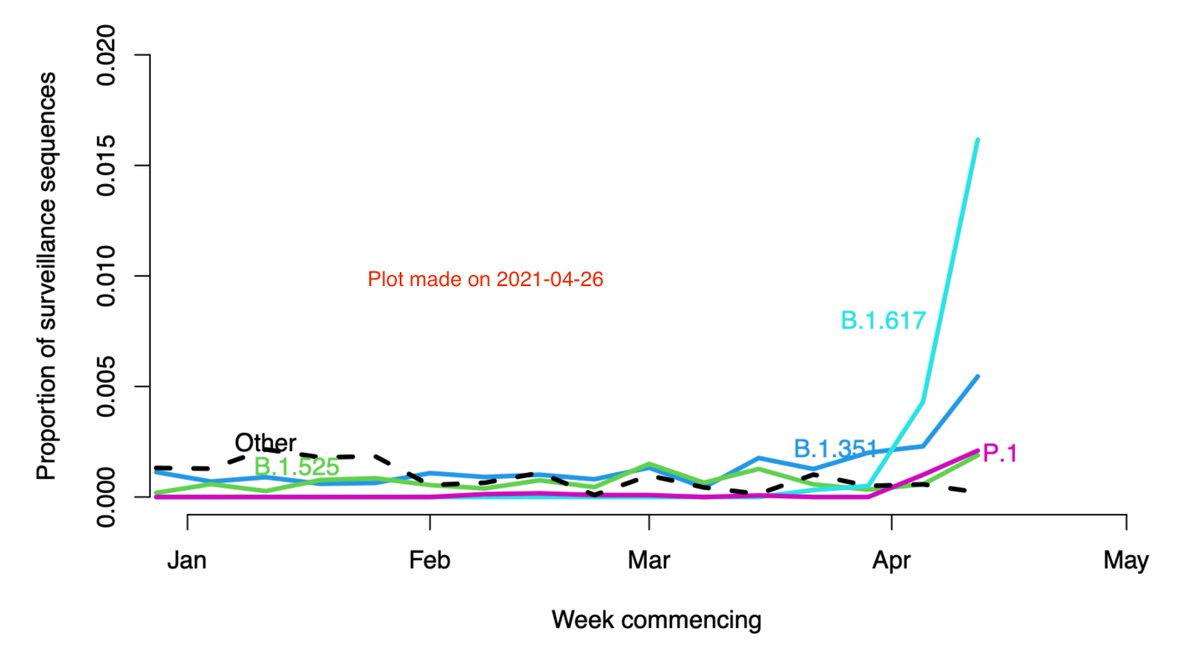

This shows the bleeding edge of what we knew about these variants as it happened. PHE designated B.1.617.2 a VOC on 7 May: four days after the last graph above, or "eventually", as the article puts it.
And getting it right mattered: reflexively designating the whole B.1.617 super-lineage would've completely confounded both real-world and lab experiments on immunity and vaccine efficacy, because it's now clear the sub-lineages have very different biological properties.
Generating data at this scale (and there is so much more than just the genome numbers which I'm most familiar with), then analysing & synthesising a very complex and (literally) evolving picture at this speed is quite simply astonishing, and unprecedented in my scientific career.
There's a heroic cast of hundreds doing this work, from @PHE_uk, the devolved Public Health agencies, and academic partners. Too many to thank individually, but I'll single out our fearless leader, @meera_chand!
• • •
Missing some Tweet in this thread? You can try to
force a refresh