A Novel and Expanding SARS-CoV-2 Variant, B.1.1.318, dominates infections in Mauritius medrxiv.org/content/10.110…
Mauritius is an example of controlling #COVID19 in Africa, with 1,700 infections and only 18 deaths. However, one variant seems to have caused the great majority of all of the infections 

Mauritius has stopped dozens of introductions of the Beta (501Y.V2/B.1.351) and Alpha (B.1.1.7) but the B.1.1.318, which has the 484K mutation (common to Beta) and the 681H mutation (common to Alpha) has caused all local transmitted cases. 
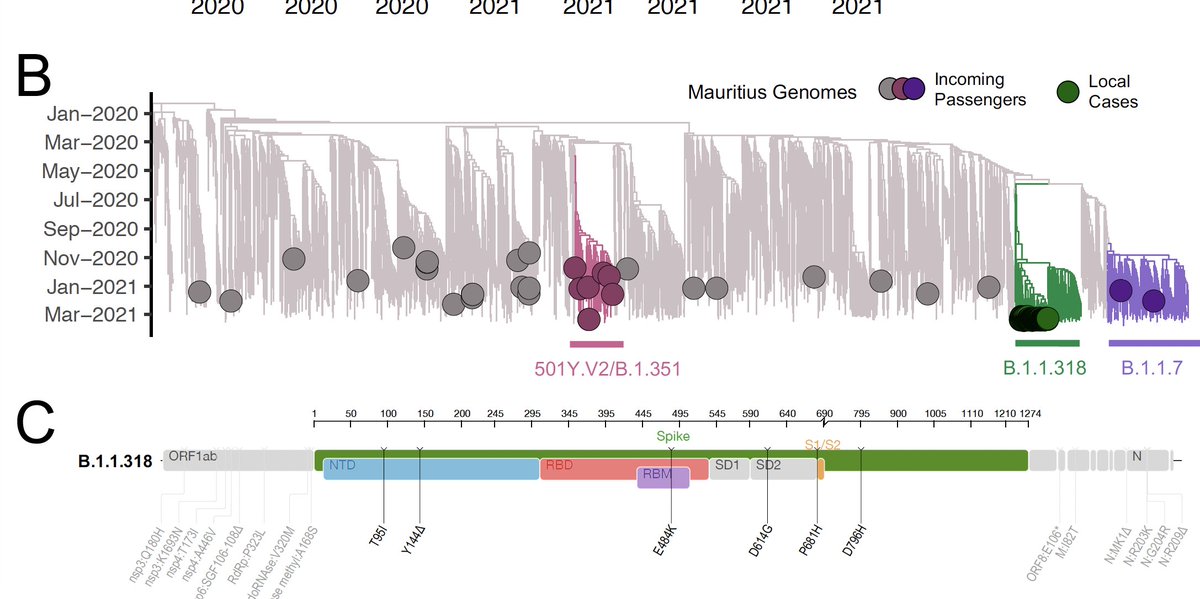
The B.1.1.318 has its origins traced to Nigeria. This variant has been highlighted as a variant of interest (VOI) in the U.K. and seems to be also transmitted at high level in Europe, which was identified as the source to introduction to Mauritius. 

This variant, B.1.1.318, is now spreading in over 35 countries and over 3,200 genomes are available at @GISAD 

Immune evasion properties introduced by Spike substitutions (e.g. E484K and Y144del) are unlikely to explain local transmission as there was presumably a low level of population immunity following a small first wave. However, it is important to detect its effect in vaccines.
At the end of May 2021, close to 20% of the population of Mauritius had received the first dose and half of those had also received the second dose of one of the vaccines.
Strong outbreak response, with widespread testing (positivity rate < 1%) seems to be able to slow the spread of this variant in Mauritius. However, public health response needs to keep focused as this variant is spreading in many other countries (>35).
Mauritius also used genomics surveillance to follow outbreaks, and in spite of a limited number of genomes, it allows the country to rank as one of the top 5 countries in genomics coverage in the world with 12% of the infections genotyped. 

In conclusion, SARS-CoV-2 genomic surveillance in Mauritius demonstrated a second wave that was dominated by a VUI, despite PCR testing and quarantine effectively preventing local transmission of other variants.
This report also highlights the need for continuous genomic surveillance to fully to understand the transmission dynamics of the VUI in Mauritius.
Pre-print open at: medrxiv.org/content/10.110…
Pre-print open at: medrxiv.org/content/10.110…
This work was done in close collaboration with the Ministry of Health and Wellness, Mauritius. The Central Health Laboratory, Victoria Hospital, Candos, Mauritius. The #franciscrickinstitute, the @nicd_sa , @krisp_news, @ceri_news and the @AfricaCDC and @WHOAFRO
Thanks to @houzhou @rjlessells @EduanWilkinson @Mittenavoig and many other colleagues from Mauritius. Together, we can expand genomics surveillance to detect and control new variants in Africa!
More information on the expanding variant in Mauritius. medrxiv.org/content/10.110…
• • •
Missing some Tweet in this thread? You can try to
force a refresh