🛠️CoVariants.org Update!🛠️
The latest push to CoVariants tweaks how sequences are classified as Variants in some cases, & you'll notice changes in some graphs as a result.
Overall, this change means better classification for lower-quality sequences👍🏻👏🏻
Read on...
1/6
The latest push to CoVariants tweaks how sequences are classified as Variants in some cases, & you'll notice changes in some graphs as a result.
Overall, this change means better classification for lower-quality sequences👍🏻👏🏻
Read on...
1/6
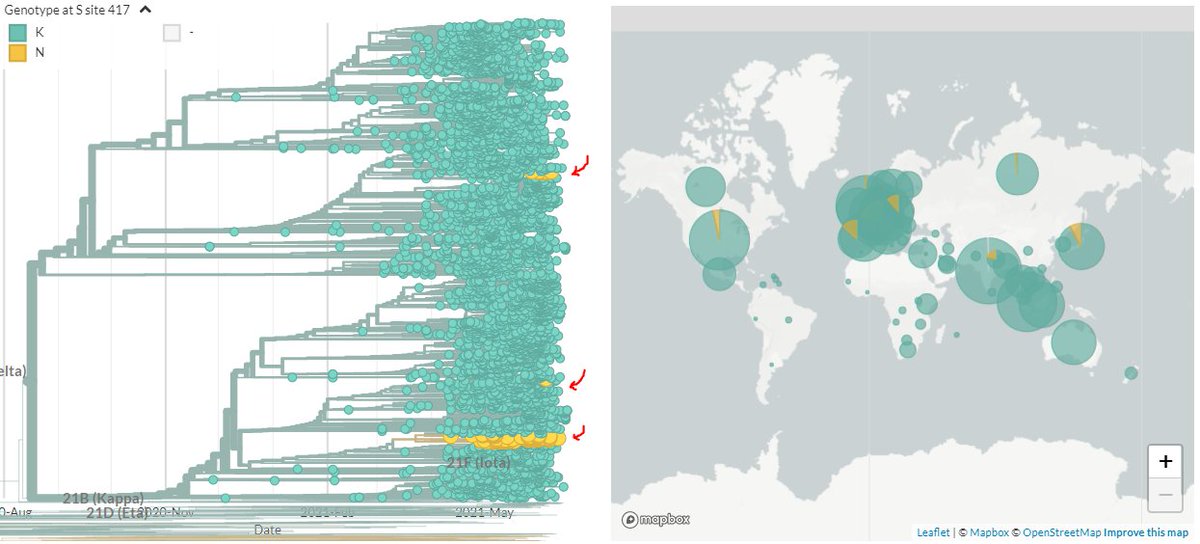
CoVariants looks at nucleotide changes to classify sequences - a sequence must have all of a list of changes to be classified.🧬🔢✅
But sometimes during a sequencing, part of the DNA might not be 'read' ("low coverage"), so we have no information for some locations. ❓
2/6
But sometimes during a sequencing, part of the DNA might not be 'read' ("low coverage"), so we have no information for some locations. ❓
2/6
If some of the positions CoVariants is looking for have low coverage & no information, they won't get classified as Variants. 🧬1⃣2⃣4⃣✖️
This can particularly impact places sequencing lower-quality sequences, as CoV will 'undercount' how many are variants! 📉
3/6
This can particularly impact places sequencing lower-quality sequences, as CoV will 'undercount' how many are variants! 📉
3/6
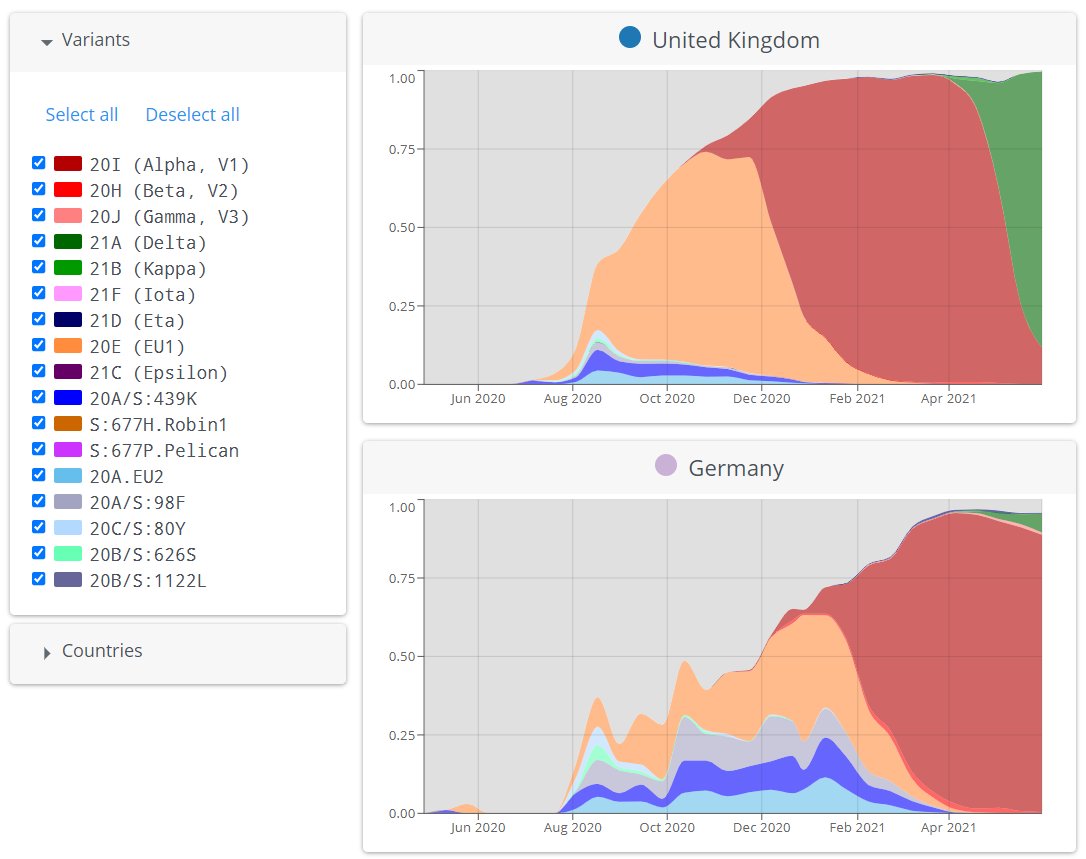
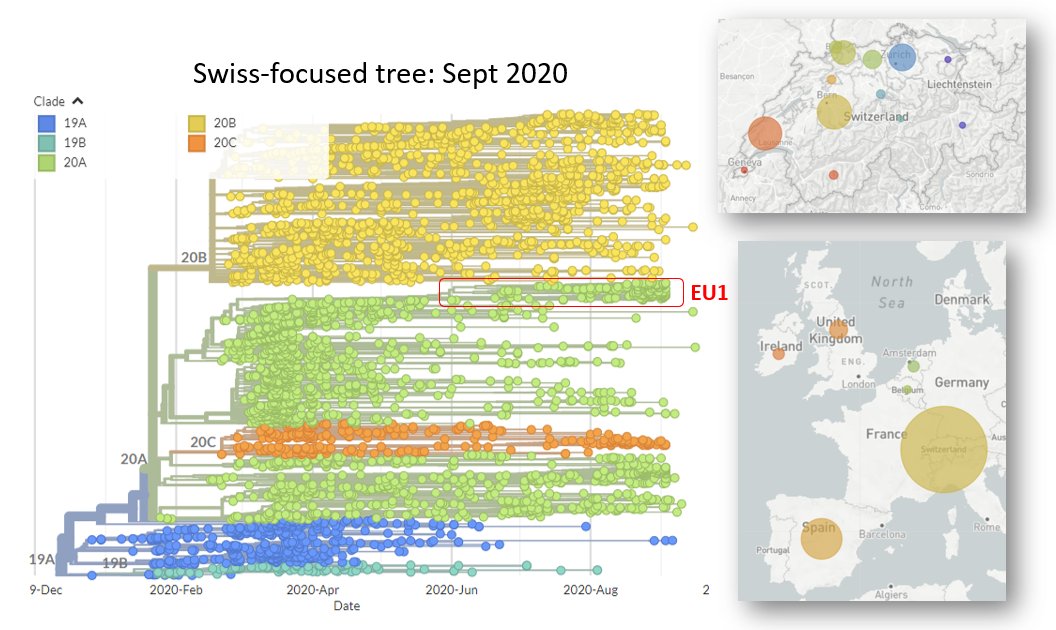
Methods like Nextclade use a more complex way to classify @nextstrain Variants, using the whole genome 📏 & phylogenetic info 🌳, which is more robust. 🔨
For Variants which are recognised Nextstrain clades, CoVariants now uses Nextclade classification! 🙌🏻
4/6
For Variants which are recognised Nextstrain clades, CoVariants now uses Nextclade classification! 🙌🏻
4/6
For most countries this doesn't make much difference in what you see in the graphs or trends, but it often does mean more sequences are correctly classified as a Variant 👌🏻✅
And in some countries it makes a big difference! See South Africa & Mexico before & after:
5/6

And in some countries it makes a big difference! See South Africa & Mexico before & after:
5/6
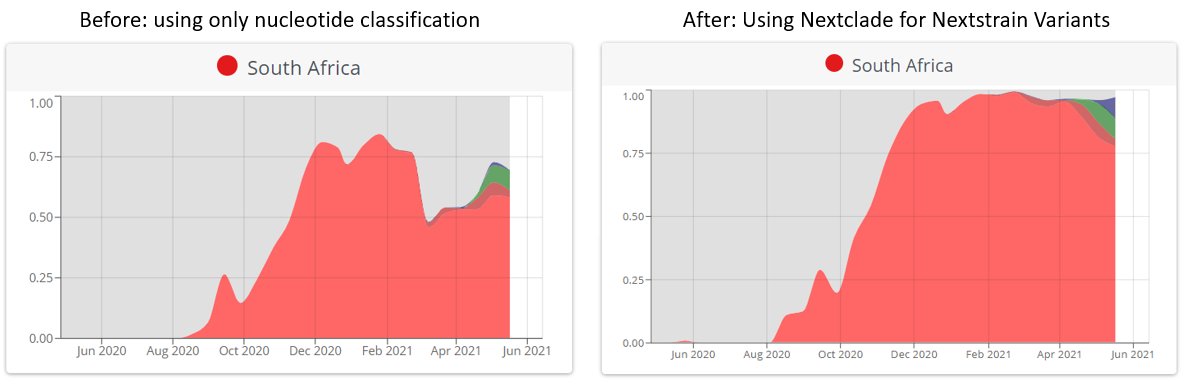

🙏🏻 Thanks to @houzhou & @Tuliodna for chatting to me about why this discrepancy was apparent in South Africa, which caused me to go take a closer look & try to resolve it! 🌍📉🔎
For the few countries that this was really impacting, I hope this is a big improvement!
6/6
For the few countries that this was really impacting, I hope this is a big improvement!
6/6
• • •
Missing some Tweet in this thread? You can try to
force a refresh