I've run preliminary @nextstrain focal builds on 20B/S:796H (B.1.1.318) (pictured) & 20A/S:210T (B.1.619).
These aren't part of CoVariants (yet?) but they've caught a couple eyes and I thought it was worth taking a closer look.
1/8
These aren't part of CoVariants (yet?) but they've caught a couple eyes and I thought it was worth taking a closer look.
1/8

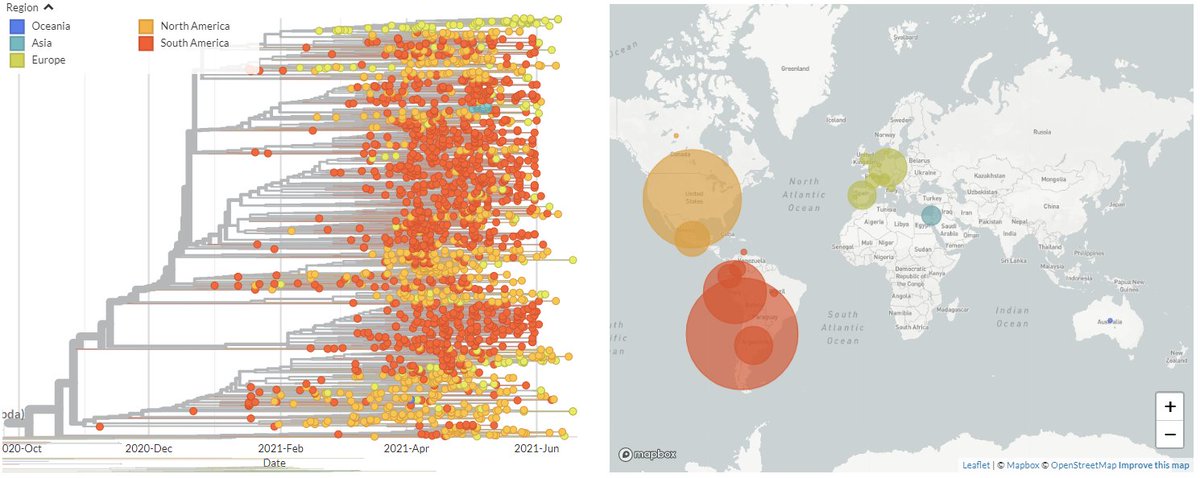
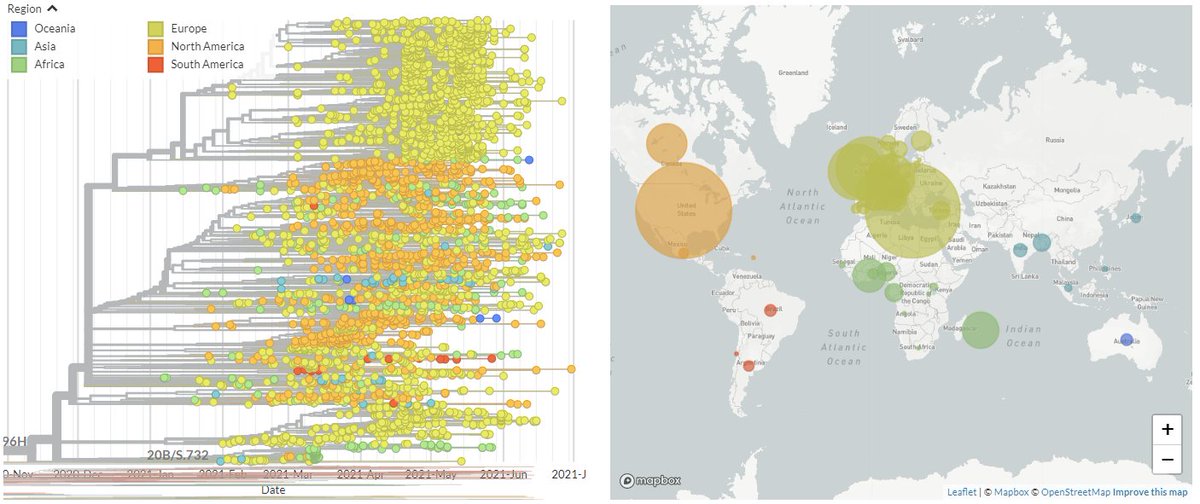
20B/S:796H (B.1.1.318) is behind most of the recent increase in 'other' variants circulating in Greece (shown in grey on the CoVariants plot, below - disregard last point, low sequence #'s).
It's very prominent in Greece, but also found worldwide.
2/8
nextstrain.org/groups/neherla…

It's very prominent in Greece, but also found worldwide.
2/8
nextstrain.org/groups/neherla…

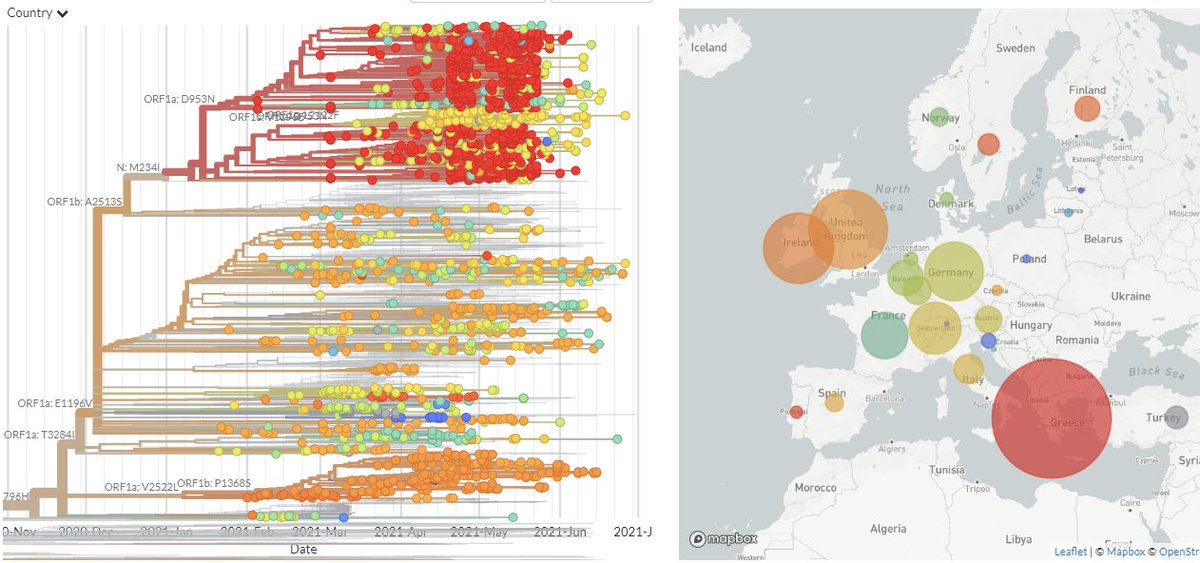
Taking a closer look at Europe only, we can see there's a large cluster in Greece, but also distinct clusters in Switzerland & Ireland, & many sequences from other countries scattered between.
3/8
nextstrain.org/groups/neherla…
3/8
nextstrain.org/groups/neherla…

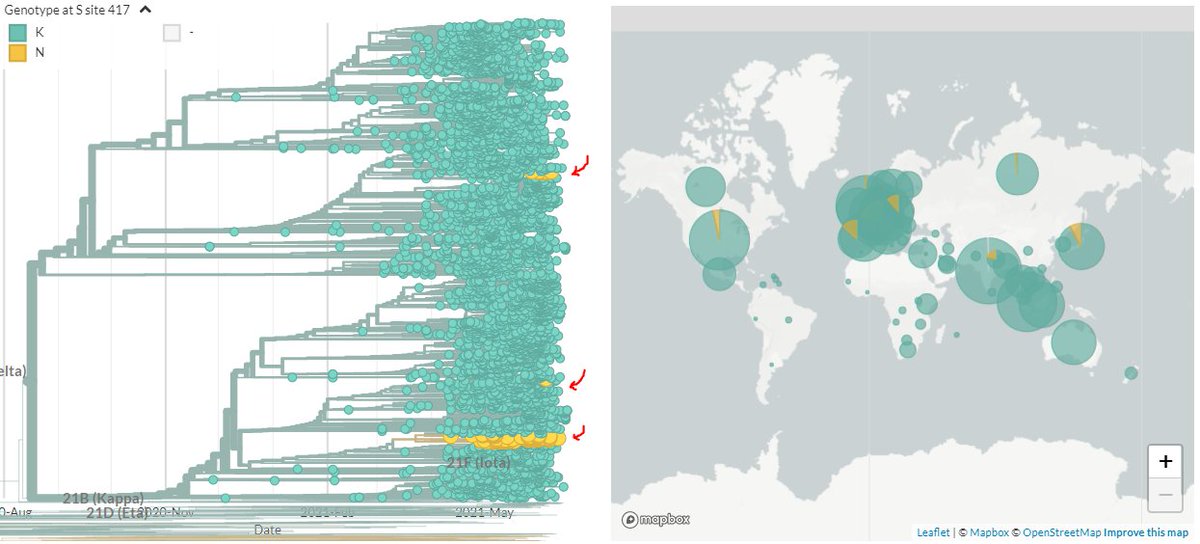
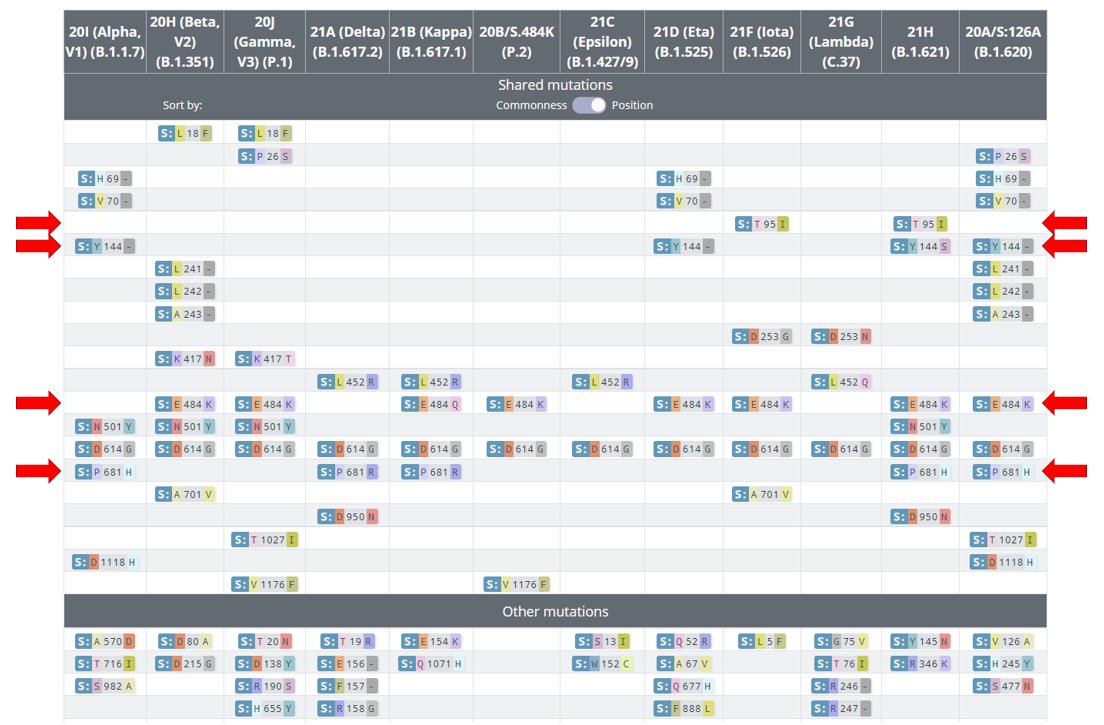
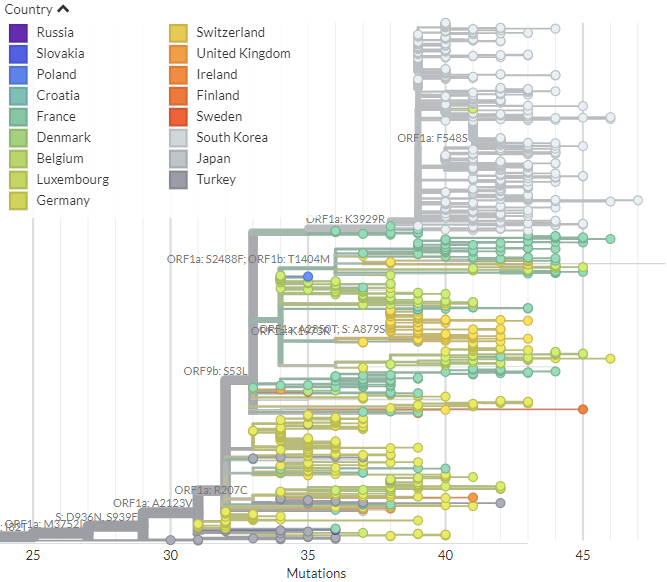
20B/S:796H (B.1.1.318) has spike mutations at S:95I, S:144-, S:484K, & S:681H which are shared with other Variants of Concern/Interest.
The distinct clusters in Switzerland, Greece, & Ireland generally don't have additional spike mutations.
4/8
covariants.org/shared-mutatio…

The distinct clusters in Switzerland, Greece, & Ireland generally don't have additional spike mutations.
4/8
covariants.org/shared-mutatio…

The fact that the significant recent cluster in Greece (or the others) doesn't have additional spike mutations suggests (but doesn't rule out) that this expansion may *not* be due to viral changes.
A reminder that it's not just the virus that determines transmission!! 🧑🏽🤝🧑🏻
5/8
A reminder that it's not just the virus that determines transmission!! 🧑🏽🤝🧑🏻
5/8
20A/S:210T (B.1.619) was initially mostly seen in Africa & Europe & seemed to be decreasing in frequency, but has recently grown again, particularly in South Korea.
6/8
nextstrain.org/groups/neherla…
6/8
nextstrain.org/groups/neherla…
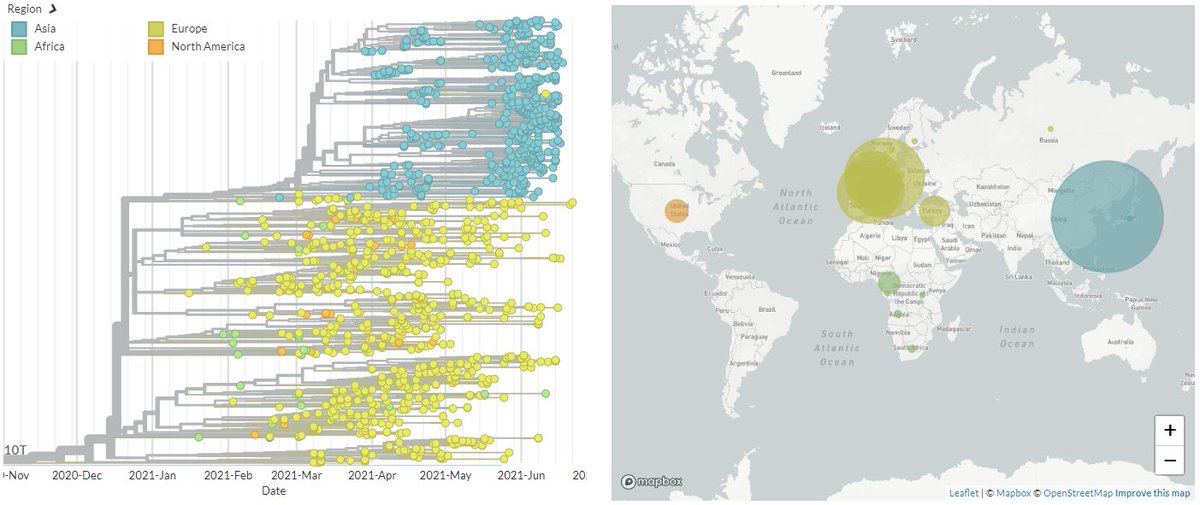
Looking just at Europe & Asia, we can see clusters in Germany, Switzerland, France, & Belgium, as well as the large South Korean cluster clearly.
7/8
nextstrain.org/groups/neherla…
7/8
nextstrain.org/groups/neherla…
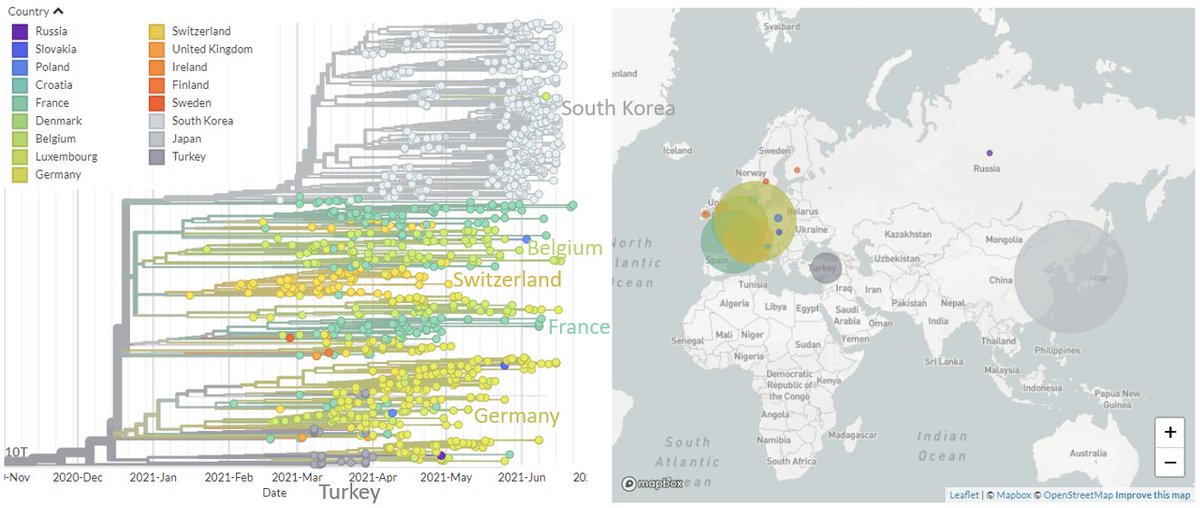
20A/S:210T (B.1.619) has a number of spike mutations, but only S:484K & S:1027I are shared with other variants of concern.
Notably, the cluster in South Korea does not have additional Spike mutations, suggesting this also may not be spreading due to viral change.
8/8

Notably, the cluster in South Korea does not have additional Spike mutations, suggesting this also may not be spreading due to viral change.
8/8

• • •
Missing some Tweet in this thread? You can try to
force a refresh