As short-read #sequencing (SRS) costs begin to drop again, undoubtedly fueled by a resurgence in competition, I suspect many liquid biopsy providers will add blood-based whole-genome sequencing (WGS) to supplement, or replace, the deep targeted sequencing paradigm.
With a few exceptions, most clinical-stage diagnostic companies build patient-specific panels by sequencing the solid tumor, then downselecting to a few dozen mutations to survey in the bloodstream.
I don't think this approach is going anywhere anytime soon.
I don't think this approach is going anywhere anytime soon.
However useful, this deep-sequencing approach suffers from several challenges:
1. It requires access to tissue.
2. It requires the construction of patient-specific PCR panels.
3. It requires significant over-sequencing ($$$).
4. It introduces a third layer of error (PCR).
1. It requires access to tissue.
2. It requires the construction of patient-specific PCR panels.
3. It requires significant over-sequencing ($$$).
4. It introduces a third layer of error (PCR).
Companies have tried to solve these issues using some combination of molecular barcoding, novel primer construction, bioinformatics, or adding in other sources of 'omics data, such as #epigenetics or DNA fragmentation patterns.
Despite rosy rhetoric, I'm not convinced any group has a durable, technical advantage on deep, targeted sequencing. To that point, the reported limits of detection (LoD) for these assays tend to hit near 0.01% or roughly 1 cancer fragment amongst 10,000 healthy fragments.
It's well established that every tumor is unique, not just from patient to patient, but from cancer type to cancer type. For example, breast and prostate cancers often have very low tumor burdens, meaning they aren't highly mutated to begin with.
Since low-burden cancers begin with fewer mutations, it makes custom panel building that much more difficult and makes the deep-targeted approach even less effective. To reiterate, we don't think the 'depth' approach is going anywhere, but it's not a panacea.
For these reasons, I was blown away last summer reading @landau_lab et al's work on 'breadth over depth'. That is, using more shallow, blood-based #WGS instead of ultra-deep, targeted sequencing. I've attached my favorite figure from the paper below.
nature.com/articles/s4159…
nature.com/articles/s4159…
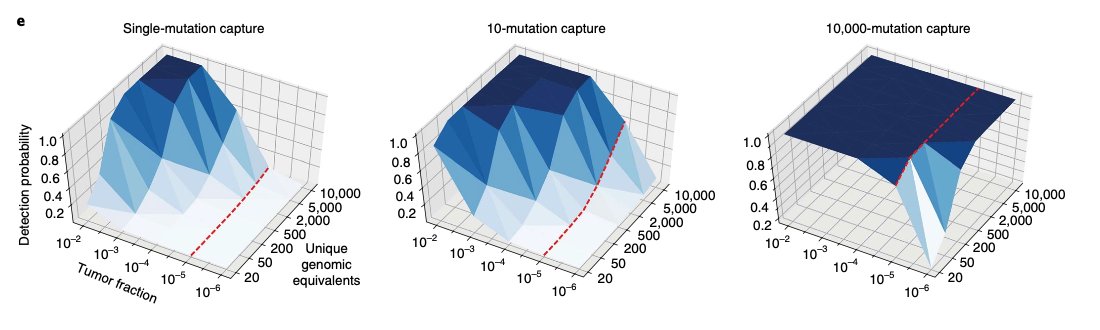
With a WGS approach, one potentially no longer needs to:
1. Get access to tissue.
2. Construct patient-specific primers.
3. Oversequence using barcodes.
4. Introduce PCR artifacts.
I'll refrain from arguing that the WGS approach will be a full-out usurper of deep targeting.
1. Get access to tissue.
2. Construct patient-specific primers.
3. Oversequence using barcodes.
4. Introduce PCR artifacts.
I'll refrain from arguing that the WGS approach will be a full-out usurper of deep targeting.
I'm sure there are unique issues and challenges here too. However, now more than ever, I'm convinced that blood-based WGS is a necessary addition for the most competitive liquid biopsy players.
Liquid biopsies are limited by the number of molecules in a tube of blood.
Liquid biopsies are limited by the number of molecules in a tube of blood.
So, it's vital that labs make the best with whatever they start with. When he was still at @freenome, @ImranSHaque gave a great presentation on the dynamics here, especially in support of a truly multi-omic approach to this problem.
ihaque.org/static/talks/2…
ihaque.org/static/talks/2…
I'll end by cautioning that implementing blood-based WGS isn't a walk in the park.
On the front-end, even though PCR errors would disappear, SRS still generates two layers of error:
1. PCR-like, on-instrument cluster generation
2. SBS-specific systematic error
On the front-end, even though PCR errors would disappear, SRS still generates two layers of error:
1. PCR-like, on-instrument cluster generation
2. SBS-specific systematic error
These aren't deal-killers, per se, but they're problematic nonetheless ~ potentially resulting in flipped base-calls and/or sequence context-specific coverage gaps.
On the back-end, there'd be gobs more data to interpret, store, and activate on behalf of patients.
On the back-end, there'd be gobs more data to interpret, store, and activate on behalf of patients.
I'm still learning about the puts-and-takes of the WGS approach and what innovations on the hardware, chemistry, and informatics sides may enable the approach and bring it (hopefully) closer to a reality.
• • •
Missing some Tweet in this thread? You can try to
force a refresh