In response to questions & comments by @hippopedoid, @adamgayoso, @akshaykagrawal et al. on "The Specious Art of Single-Cell Genomics", Tara Chari & I have posted an update with some new results. Tl;dr: definitely time to stop making t-SNE & UMAP plots.🧵biorxiv.org/content/10.110…
In a previous thread I talked about the (von Neumann) elephant in the dimension reduction room: t-SNE & UMAP don't preserve local or global structure, they distort distances, and they are arbitrary. Almost everybody knows this but they are used anyway...
https://twitter.com/lpachter/status/1431325969411821572?s=20
There were some interesting technical questions about our work. One question was the extent to which PCA pre-conditioning affects results. We examined this (Supp. Fig. 3). Tl;dr: it's time to stop making t-SNE & UMAP plots (with or without PCA pre-conditioning). 

Several people asked whether UMAP preserves neighbors. We didn't initially report on this because others have done so previously, and found that UMAP does *not* do a good job preserving them. @hippopedoid et al. showed this in nature.com/articles/s4146… 

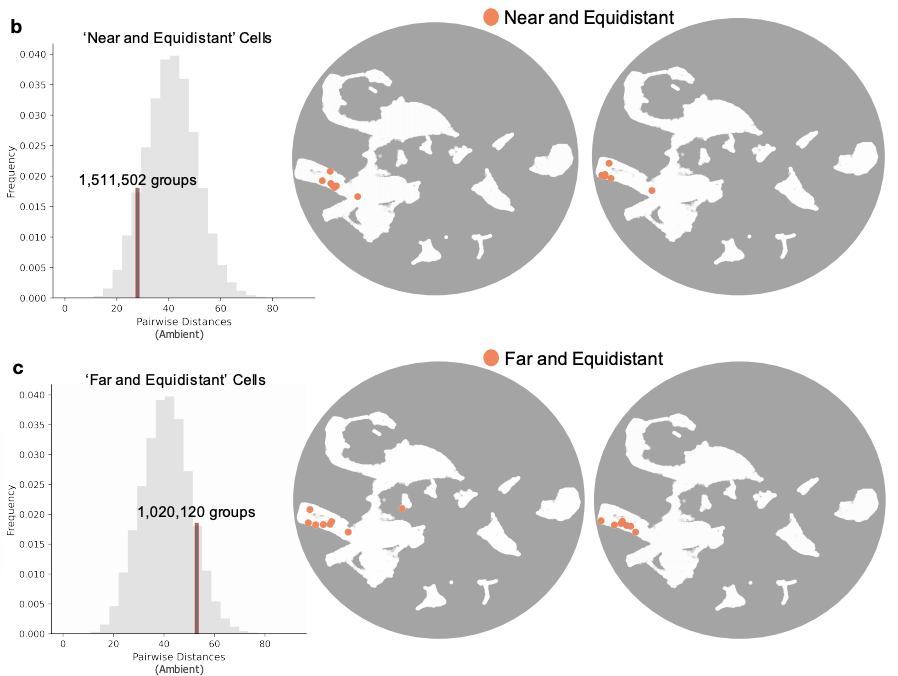
But since people ignored them, we added neighbor results for our data (Supp. Fig. 7). Tl;dr: t-SNE & UMAP scramble neighbors and do not preserve local structure. It's true that PCA is worse at preserving local structure but rotten apples aren't tastier than longer rotting apples. 

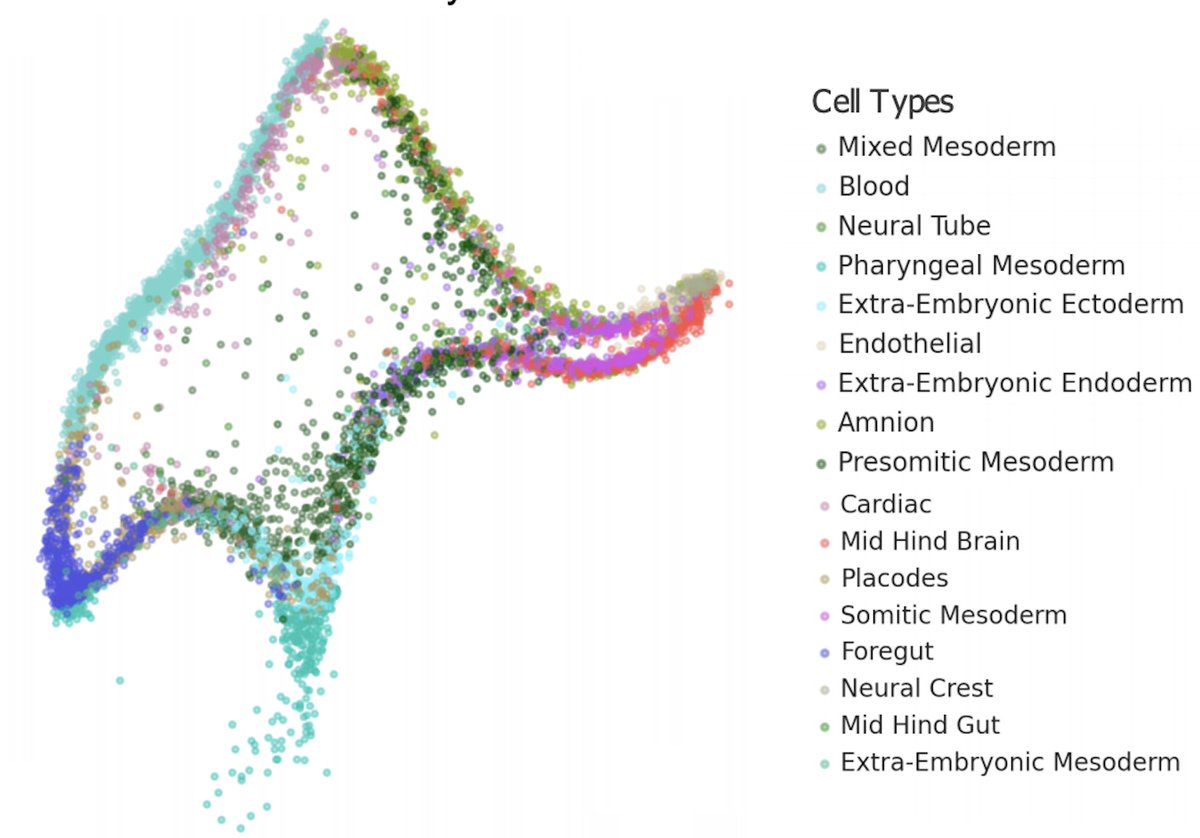
Some people argued that UMAP on MNIST shows it is a useful visualization tool. So we looked at MNIST carefully. UMAP is quantitatively terrible and visually misleading for an analysis of MNIST. It seems good in @hippopedoid's tweet but that's a mirage.
https://twitter.com/hippopedoid/status/1437421945956470785?s=20
First of all, the digits are mixed up in some of the seemingly "pure" clusters. The illusion happens because points are plotted over. Re-ordering the way in which points are displayed shows that many digits are misplaced. This is borne out in a quantitative analysis of the data. 

These bad visuals don't reflect the intrinsic structure of the MNIST data. In fact, it's possible to perform digit assignment very accurately (99.87%) with the MNIST data. It just requires operating in dimension much greater than 2. paperswithcode.com/sota/image-cla…
t-SNE and UMAP also scramble the relative cluster placements. Clusters that are far apart appear near and vice versa. The sizes of the clusters have no relation to their actual sizes. 

A UMAP of MNIST can even fail to reveal the correct number of digits, as this UMAP made by @akshaykagrawal shows (from his @GoogleColab notebook made to try Picasso). In it, there are two distinct clusters for the digit 8, with one sandwiched between 3 and 4. 

It's not that two-dimensional visualizations are always useless. It's just that t-SNE and UMAP, when applied to high-dimensional data, provide visualizations that require users to navigate non-biologically interpretable parameters and tune visuals to their expectations.
It's convenient to say "these methods don't try to preserve distances", but the way embeddings are used inherently assumes distances and ordinal relationships have been preserved. In biology entire methods depend on such assumptions (e.g., Monocle3, velocyto, scvelo, etc.)
As a result of a lot of handwaving, hyperbole and hype, there also seems to be tons of confusion about what UMAP & t-SNE do, including confounding between their loss functions, and the properties the embeddings they produce actually have.
For example, some people pointed out that t-SNE and UMAP weren't meant to preserve local and global structure... only distance.
https://twitter.com/verwer/status/1431332940680630272?s=20
Others disagreed with that, and noted that t-SNE and UMAP were supposed to preserve *some* global structure (quoting, for example, the authors of t-SNE):
https://twitter.com/juanbuhler/status/1431350193291620352?s=20
In disagreement with that, some noted that t-SNE and UMAP were not aimed at preserving global structure, but rather were preserving *local* structure. This is a common belief (based on the loss function), but as we and others have shown, it's just not true.
https://twitter.com/TAH_Sci/status/1433451046295179271?s=20
One person pointed out UMAP and related methods "don't try to preserve [distances]" but instead "put similar items near and dissimilar items not near [each other]". 🤔
https://twitter.com/akshaykagrawal/status/1431678631718227971?s=20
The confusion about UMAP and t-SNE is not the fault of users. It reflects lack of theorems about their performance, hype by some, and the fact that the images they make with thousands or even millions of points can be beautiful (even if misleading).
https://twitter.com/hippopedoid/status/1318917878364672001
The upshot: there's no need to throw a UMAP monkey wrench into your analysis. We can't help but make inferences when we look at these visuals. But "Specious Art" shows they are misleading, and they are also arbitrary (Picasso image by @sinabooeshaghi github.com/sbooeshaghi/pi…). 

• • •
Missing some Tweet in this thread? You can try to
force a refresh