Our latest study is about an immunocompromised patient with persistent COVID ➡️ treated with remdesivir but developed resistant mutation ➡️ was then cured by monoclonal Ab cocktail. Study by @gandhisk @sneakyvirus1 @epidememeology @marioph13 et al. (1/)
medrxiv.org/content/10.110…
medrxiv.org/content/10.110…
This patient received a course of rituximab (B cell depleting Ab) and bendamustine (chemo agent) for the treatment of Stage IV Non-Hodgkin's lymphoma. As a result, patient had extremely low number of T cells and low lymphocyte counts overall. Analysis by @peowenlu (2/) 
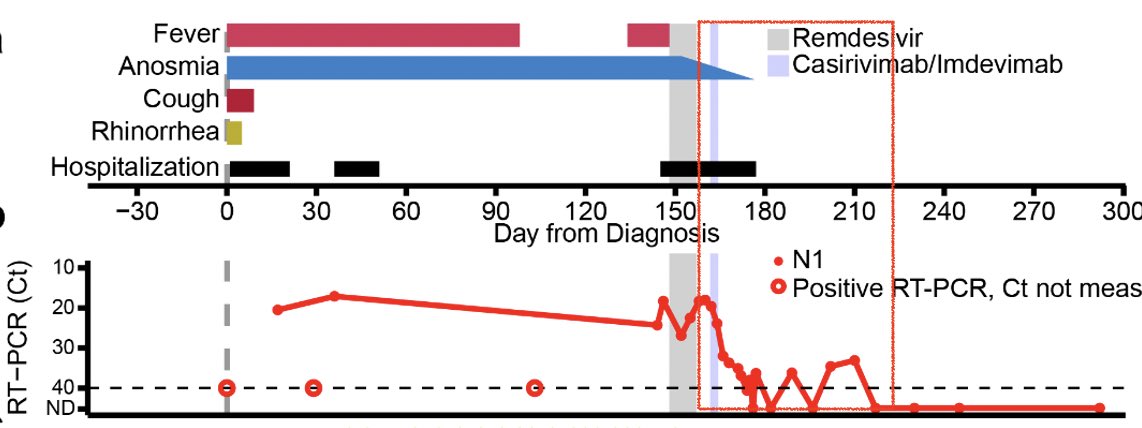
So when she was infected with SARS-CoV-2, she was unable to clear the virus for over 150 days. She developed persistent fever, anosmia and ground glass opacities in her lungs. Her viral load stayed high until the remdesivir treatment. (3/) 


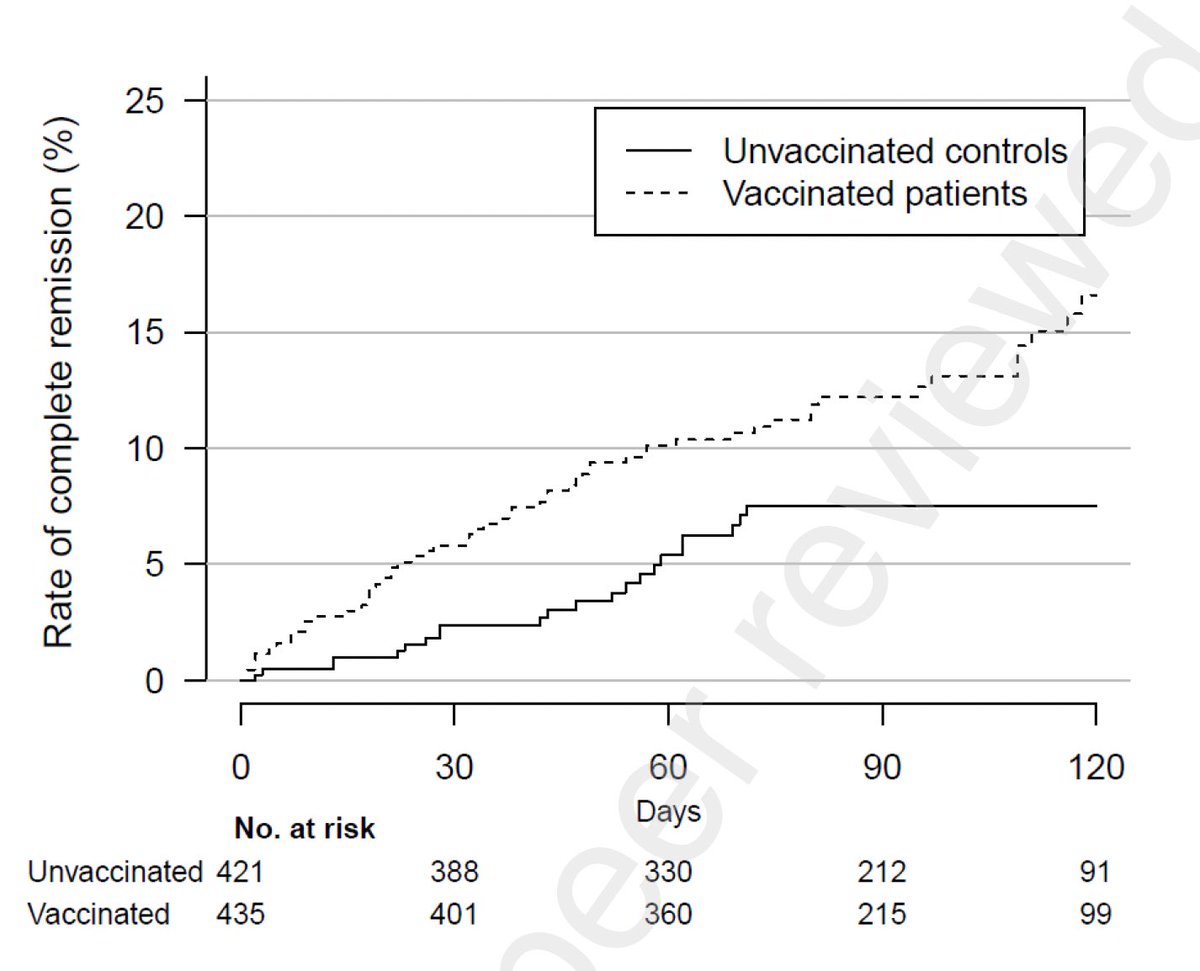
Remdesivir initially brought her viral load down but this did not last for long. Viral load came up during the course of remdesivir treatment (see shaded gray timeline). (4/) 

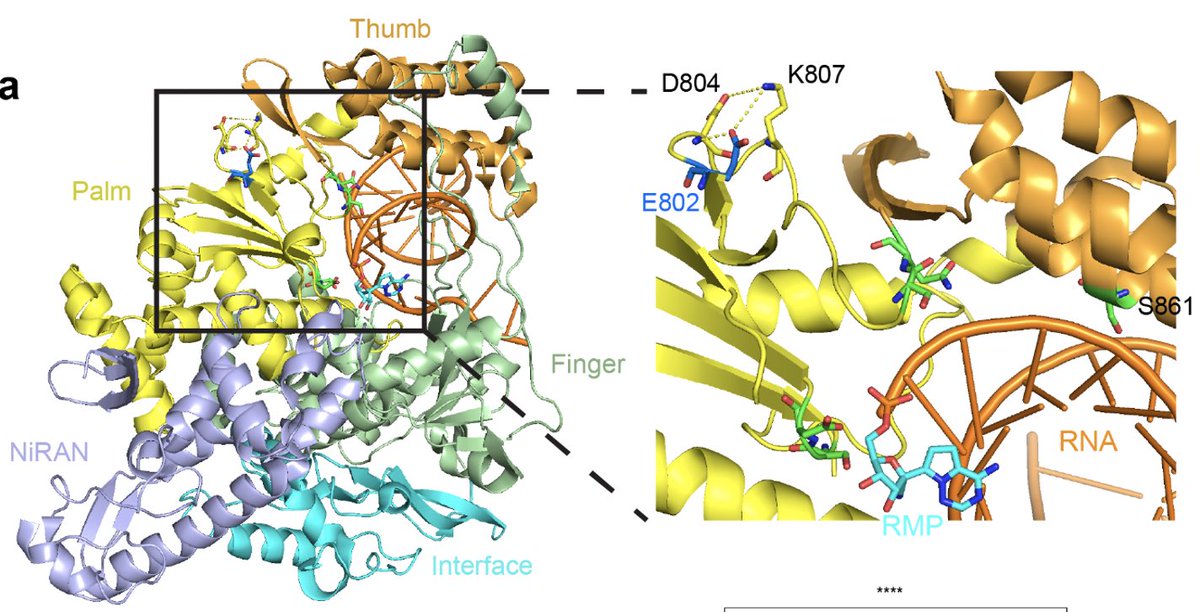
Why did the virus rebound during the remdesivir treatment? To get at this question, genome of the virus was sequenced. We found an intriguing mutation in the nsp12 gene, E802D, that appeared during the drug treatment. (5/) 
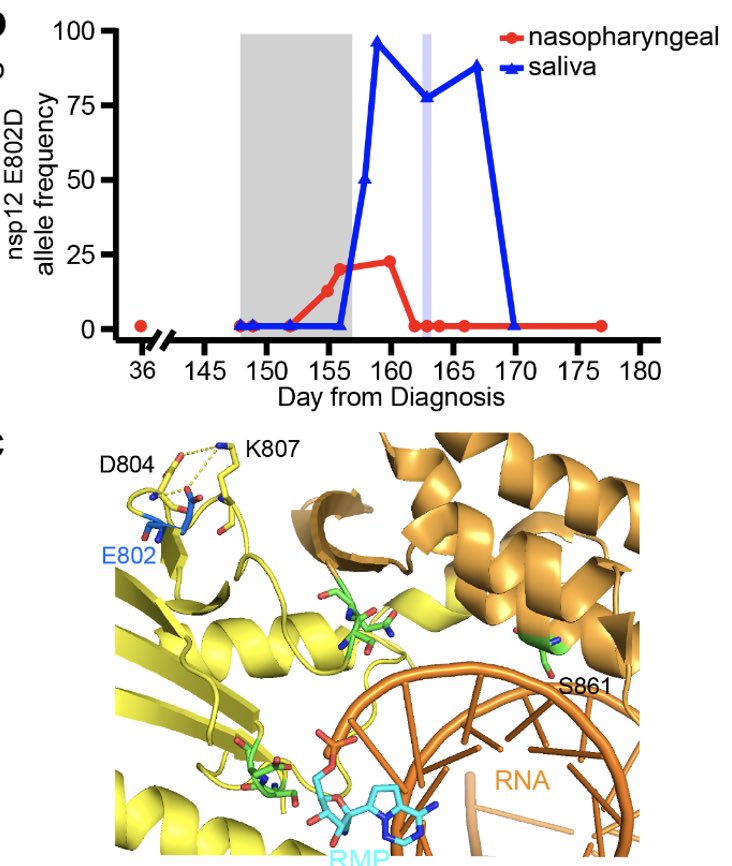
Nsp12 is a key component of the RNA-dependent RNA polymerase responsible for viral replication and transcription. Fortunately for us, a prior study already found that nsp12 E802D confers resistance to remdesivir in an in vitro selection experiment. (6/)
journals.plos.org/plospathogens/…
journals.plos.org/plospathogens/…
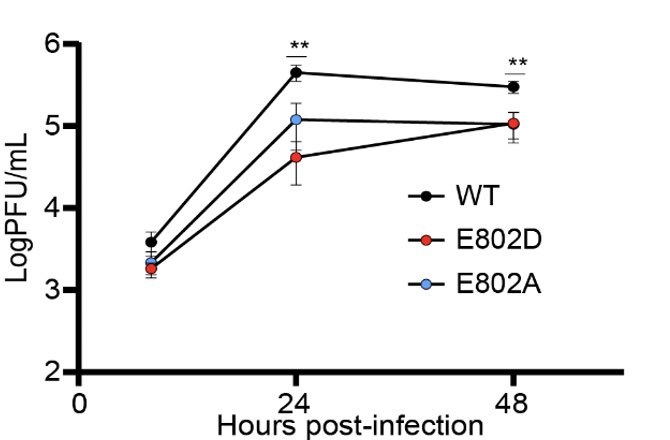
Since the genome of the patient’s virus contained many other mutations, we needed to know if the nsp12 E802D mutation was sufficient to confer remdesivir resistance. @Marioph13 heroically synthesized E802D mutant and showed that it’s indeed the case. (7/) 

We speculate that the nsp12 E802D mutation distorts the active site in a way that either 1) enables it to exclude remdesivir or 2) alleviates the steric clash mediated by S861, bypassing remdesivir-mediated chain termination. Insights from Anna Pyle and Wenshuai Wang 👇🏽 (8/) 
How frequent is the nsp12 E802D or any other substitutions at 802? Luckily very rare. Only 122 (0.003%) and 270 (0.007%), respectively, of the 4.1M genome sequences obtained from patient isolates in the @GISAID database have such mutations. (9/)
This is likely because introducing these mutations to the nsp12 make the virus less fit to replicate in the absence of remdesivir. (10/) 
Since the patient was unable to control the virus, the doctors gave her a monoclonal antibody cocktail, casirivimab/imdevimab. Remarkably, her viral titers went down, along with anosmia! Interesting correlation btw clearance of persistent virus and gain of sense of smell 🤔 (11/) 
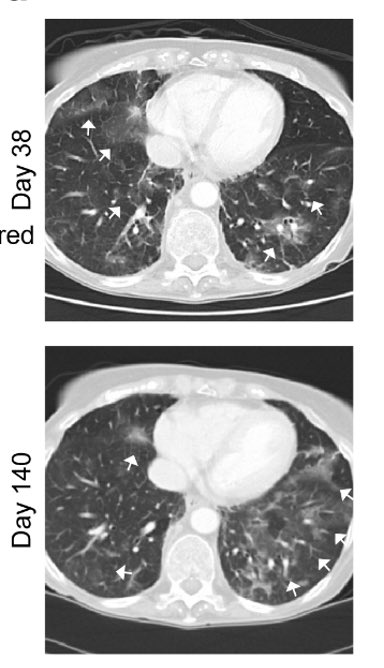
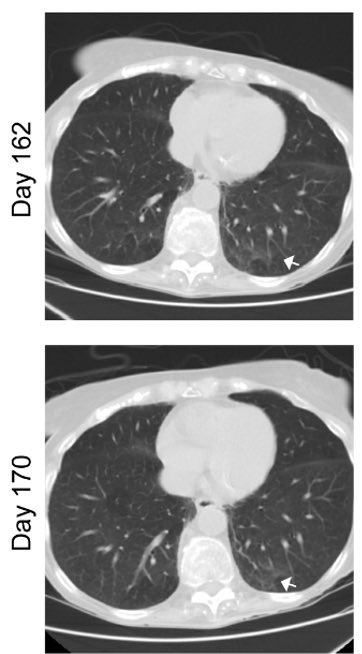
With the clearance of the virus, ground glass opacities also disappeared, along with diminished inflammatory signatures. (12/) 

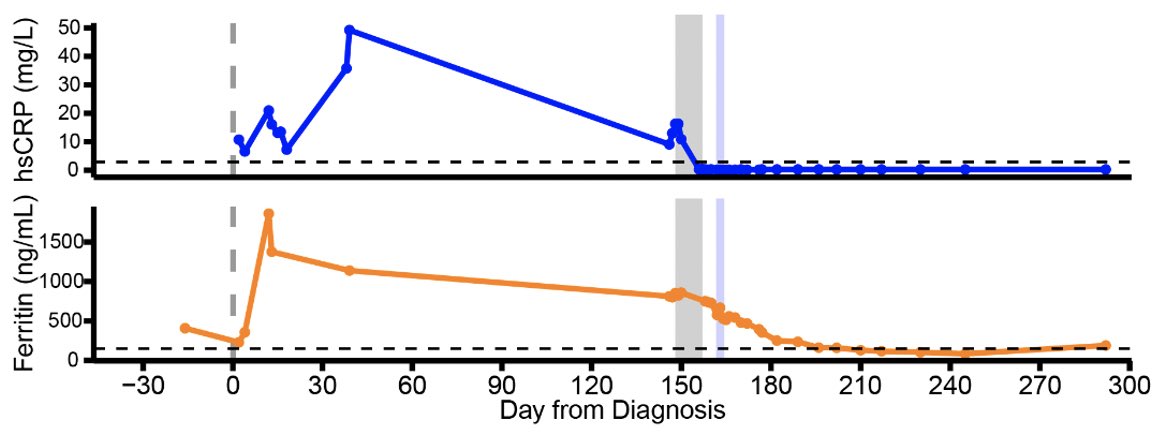
This study illustrates that in this immunocompromised person, endogenous immune responses were unable to control the virus, and that remdesivir resistance arose while on treatment. Monoclonal Abs were effective in clearing persistent virus and restoring sense of smell. (13/)
Another point to highlight is that other than fever and anosmia, this patient was able to tolerate high viral burden for months. Perhaps the impaired adaptive immunity contributed to a disease tolerance phenotype? (14/)
As always, this was a heroic effort by many. In addition to the 4 lead authors, many others contributed incl @peowenlu @WilenLab @GreningerLab @wade_schulz and Albert Ko. Special thank you to the members of @NathanGrubaugh for help and advice 🙏🏼 (end) 

• • •
Missing some Tweet in this thread? You can try to
force a refresh