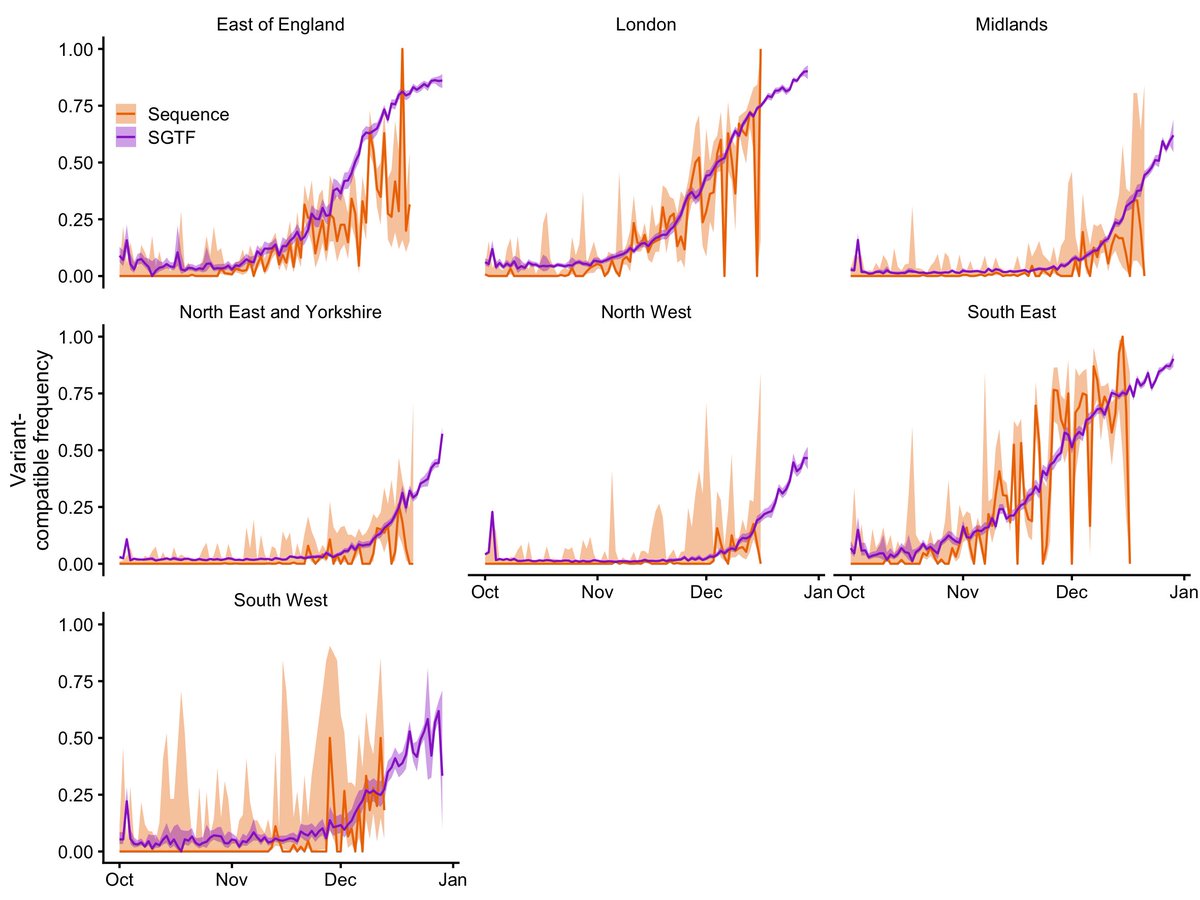
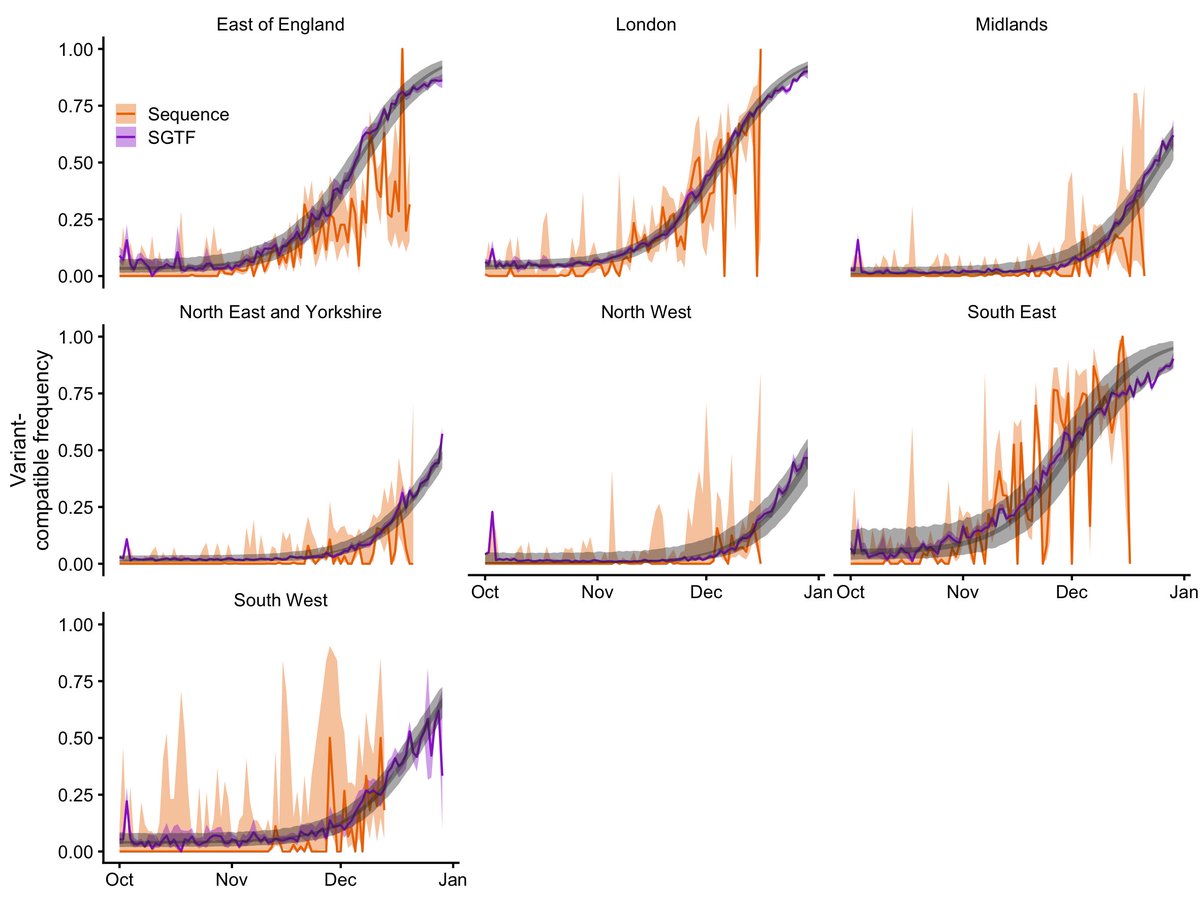
COVID-19 surveillance, England. The most recent 5 days (to 28 Nov) show an increase in S gene target failure in community testing data. An explainer follows. 🧵 

But before I go on, I will remind my scientific and media readers that SGTF is only a proxy for Omicron, and is also found in other SARS-CoV-2 lineages. Lineage identity can only be confirmed by sequencing, and longer-term surveillance is required to establish any trend.
I should also add, the fact that there has been an increase in SGTF isn't necessarily surprising — we have 22 confirmed Omicron cases in England as of today, so there was going to be an SGTF signal sooner or later. This isn't meant to be shocking news. Still, good to be aware.
Anyway. About half of community PCR tests in England are processed by labs that use a commercial detection kit that looks for three coronavirus gene targets: S, N, and ORF1ab. In the Omicron variant, the S (spike) gene has mutated such that these kits can only detect N & ORF1ab.
This is because these kits recognise specific gene fragments, so the test can't detect coronavirus genes if there is a mismatch between the primer sequence from the kit and the genetic material being analysed.
When the lab only detects N and ORF1ab, the sample is still positive for SARS-CoV-2, but the S target hasn't been detected. This is known as S gene target failure or SGTF. The Alpha variant from last year also had S mutations that caused SGTF.
We can only confirm which variant a swab has picked up by sequencing the virus's genome, but that takes time and resources. So when a new variant has SGTF, this can serve as an early signal that there may be cases of the new variant that haven't been confirmed via sequencing yet.
It's not a perfect signal. Some rare lineages carry SGTF without being a new variant, and SGTF can sometimes occur just because the viral concentration on a swab is low. So in the plot above, the low level of SGTF between August and mid-November is just background noise.
But in the last five days (24 – 28 Nov), the level of SGTF has gone up from its usual ~0.1% to ~0.3%. These are not huge numbers of cases — this represents about 60 more SGTF cases than we would expect to see in these data given the background prevalence of ~0.1%.
However, this number will probably go up, as the last 2-3 days of data are still filtering in — here's the total number of cases for England over the last few weeks for which we can determine SGTF presence or absence. 

Given that Omicron causes SGTF, while the otherwise globally dominant Delta variant doesn't, these "excess" SGTFs are most likely Omicron cases, at least some of which have yet to be confirmed via sequencing.
When it looks like a new variant is increasing in frequency, the main concern is that this could indicate community transmission. But this isn't *necessarily* what these data are showing at the moment.
We're also probably importing more Omicron cases from abroad as time goes on, since it is spreading to other countries and not all cases can be stopped by the border measures we have in place.
So, scientists in England will be watching this data stream carefully over the next several days and weeks to work out what is happening.
Incidentally, these SGTF cases are concentrated in similar areas of England to the Omicron cases that have been confirmed so far. This graphic from the Guardian illustrates the spatial pattern well. 

In a manner of speaking, we have been lucky in the UK that first Alpha had SGTF, then Delta didn't, and now Omicron does. Each time, we have been able to use the presence or absence of SGTF to detect probable VOC cases a few days before the sequencing data has been available.
You can see this in the graph in the first tweet — the decrease in SGTF through mid-July is Delta (no SGTF) replacing the previously dominant variant, Alpha (SGTF).
The SGTF signal also makes it easier to monitor the severity of a new variant, since it gives us another way to classify a case's lineage and then to see whether a given lineage is more or less likely to lead to severe disease.
• • •
Missing some Tweet in this thread? You can try to
force a refresh