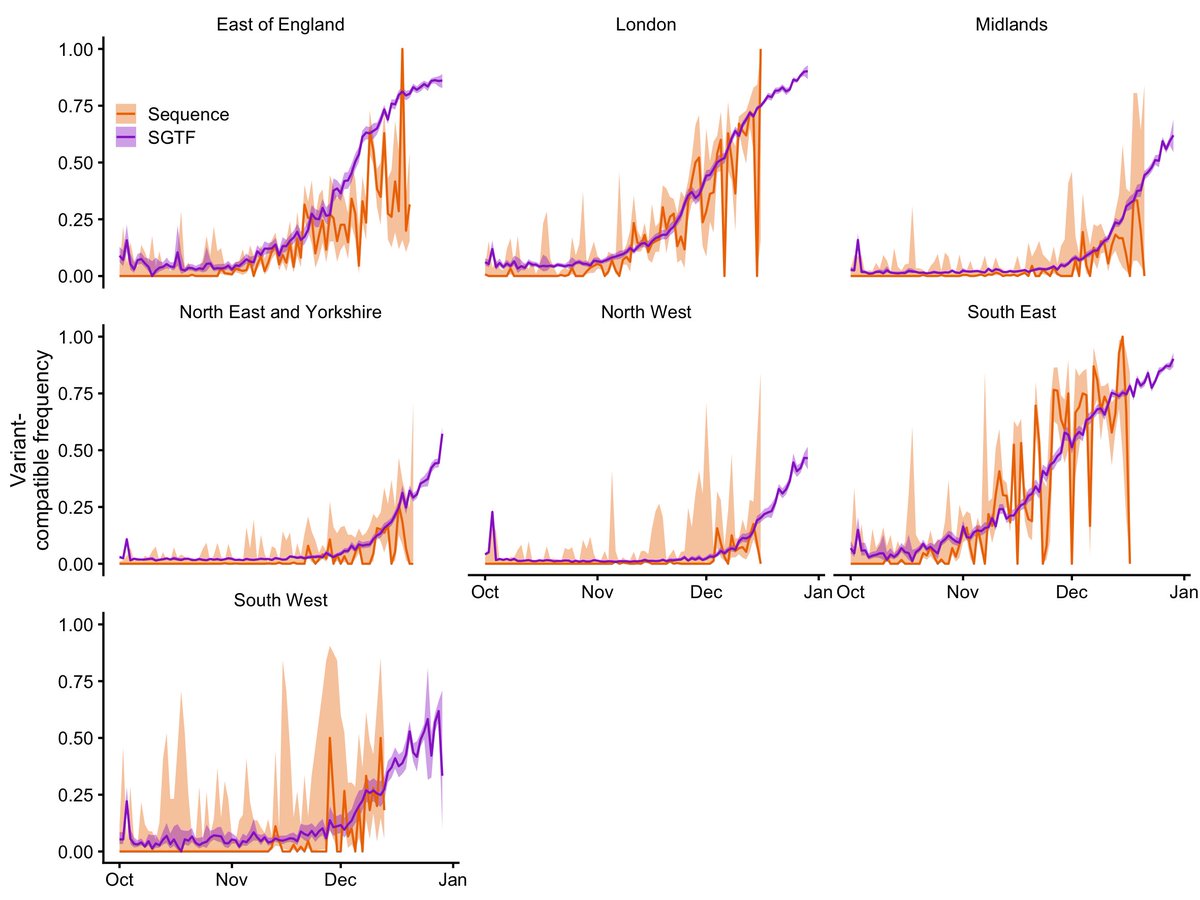
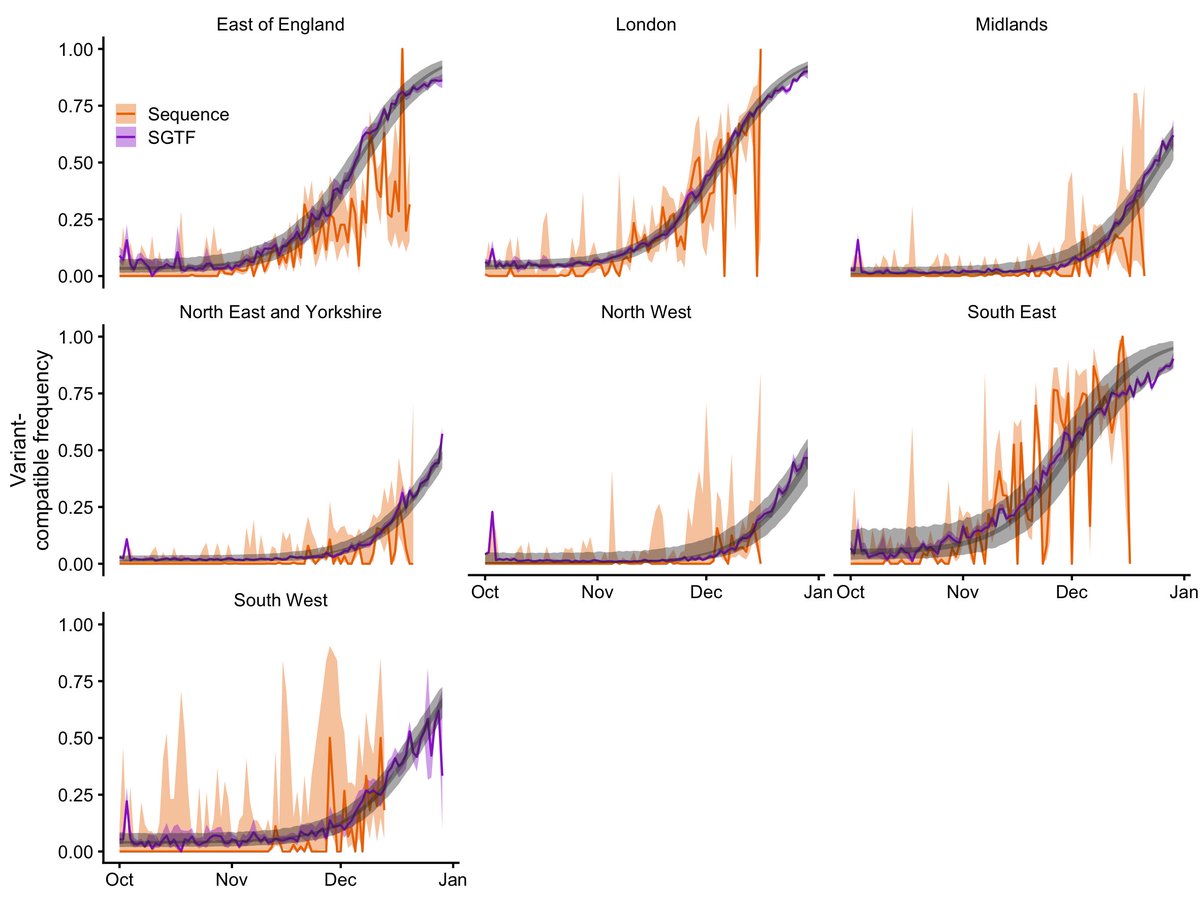
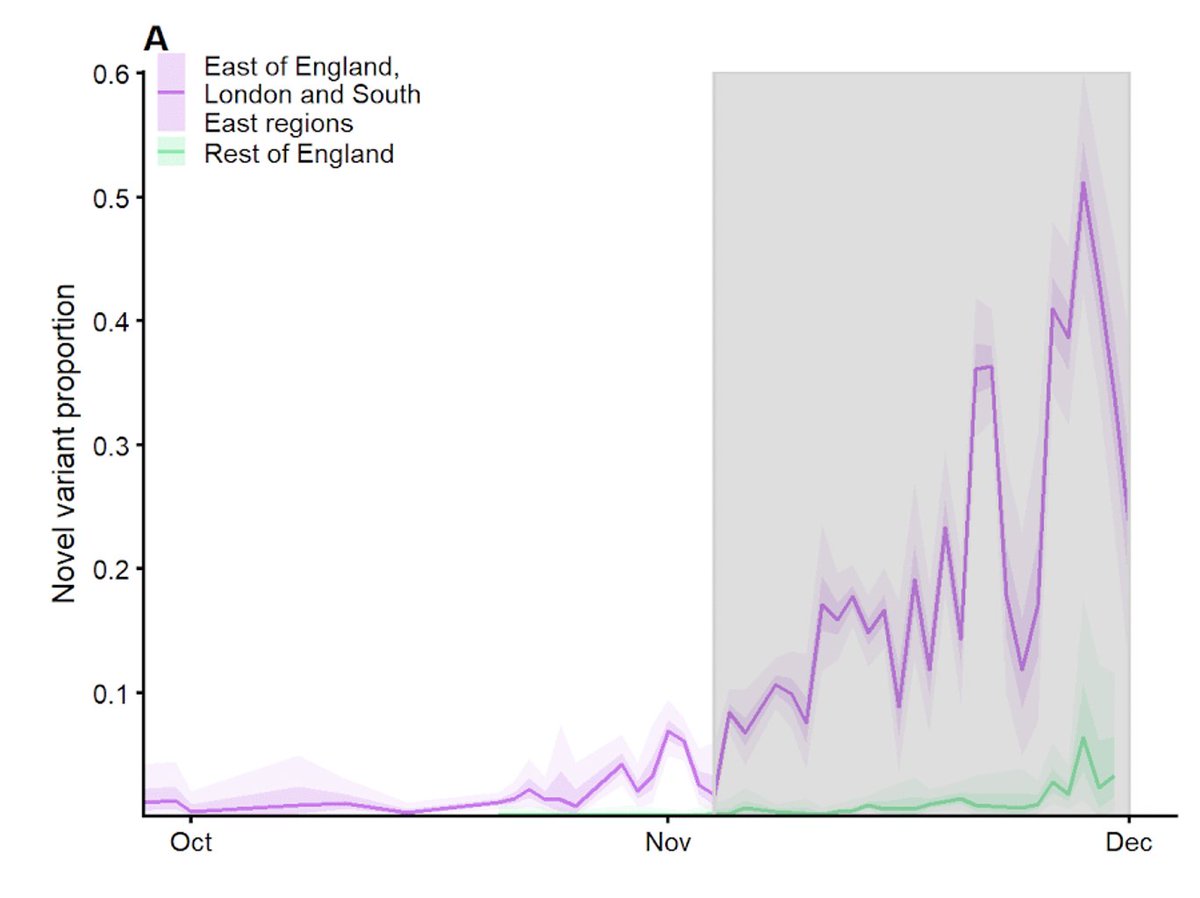
Our paper “Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7” is in Nature today. We find that B.1.1.7, the UK coronavirus variant identified in late 2020, is associated with 55% higher COVID-19 mortality than other lineages. nature.com/articles/s4158… 1/16 

The LSHTM team has been analysing B.1.1.7 for signs of increased or decreased severity since late December. We first identified a signal of higher mortality in mid-January. This led to an announcement by the PM and @uksciencechief on 22 January. bbc.co.uk/news/health-55… 2/16
In that early report, we estimated that B.1.1.7 was associated with a ~35% increase in mortality, based on around 2600 deaths, of which 384 had the SGTF marker for B.1.1.7. Our updated analysis is based on 4900 deaths, around 3100 of which had SGTF. 3/16 

Using a statistical technique called a Cox proportional hazards model, we found that people infected with B.1.1.7 had a 55% (95% CI 39–72%) higher rate of mortality within 28 days of receiving a positive test. 4/16 

The analysis controls for age, sex, deprivation, ethnicity & care home/residential/other residence type. We only compare death rates among people living in the same local authority, who were tested on the same day, to control for changes in test rates and hospital pressure. 5/16 
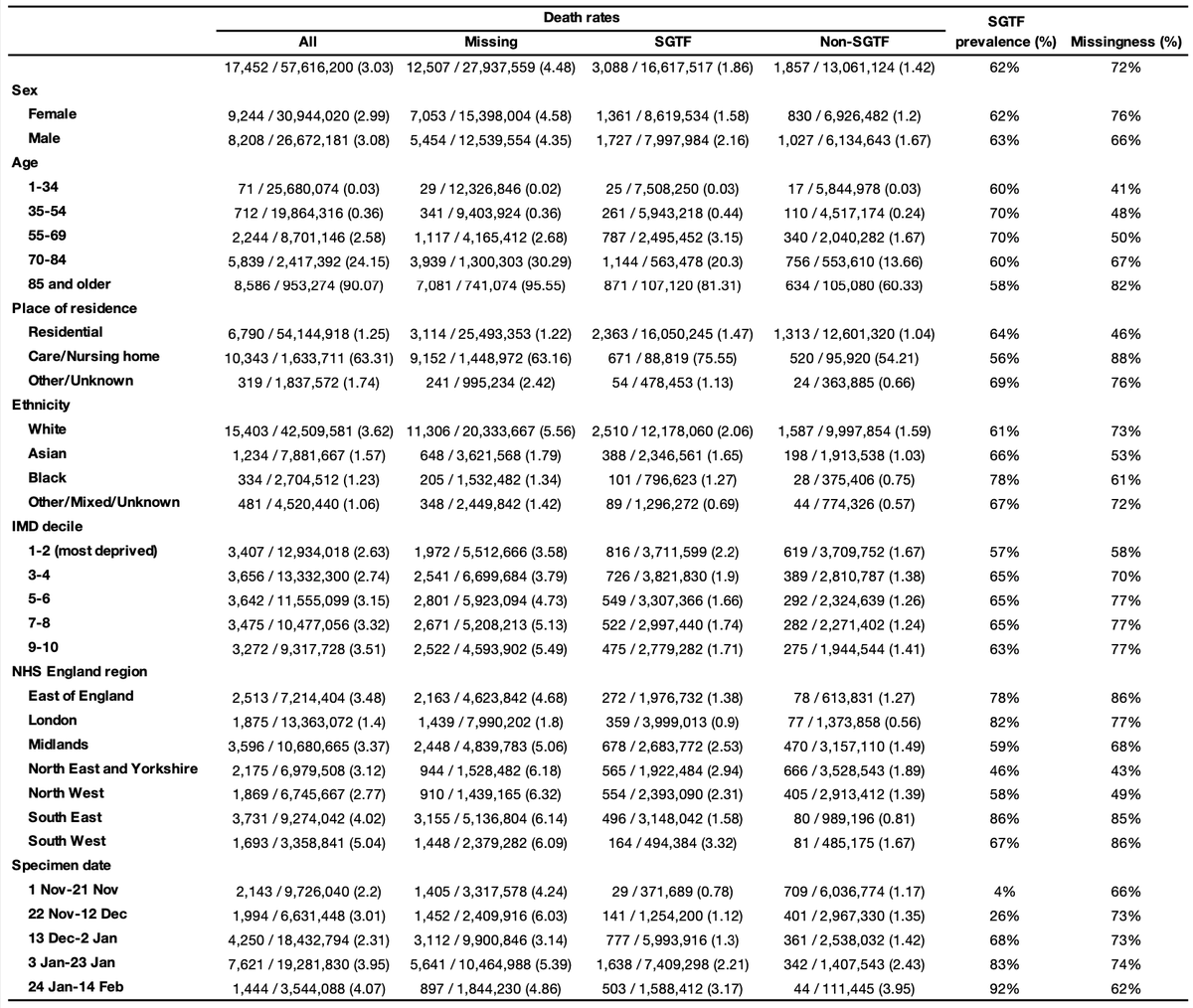
We did not find any statistically significant differences in the increased mortality rate associated with B.1.1.7 by age, sex, IMD, ethnicity, or residence type. 6/16 

While we use a statistical model to account for all the confounding factors, you do not need a model to see that B.1.1.7 is associated with higher mortality in this study sample. The raw data show the pattern clearly. The stats are there to get a clearer estimate. 8/16 

I think the most convincing way of looking at the raw data is by comparing the observed number of deaths per follow-up day, stratified here by a number of characteristics. Diamonds higher than circles = increased mortality among subjects with SGTF (the marker for B.1.1.7). 9/16 
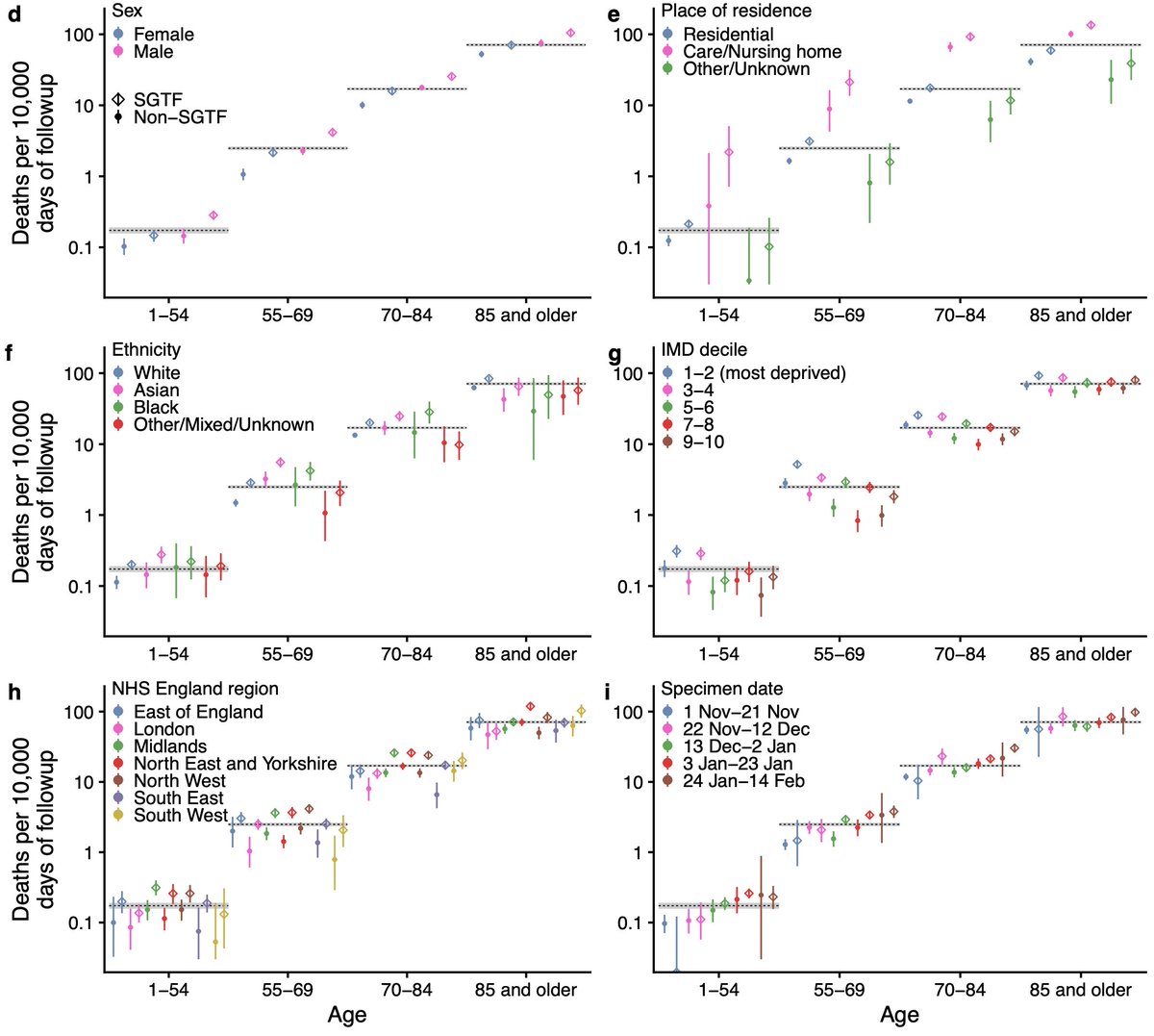
Our study sample is people tested in the community. If those infected with B.1.1.7 were less likely to get a test, we might only detect severe cases & hence overestimate mortality. But we found no major differences in test seeking (if anything, slight increase for B.1.1.7). 10/16 

Other groups working contemporaneously and since have found similar results:
Challen et al bmj.com/content/372/bm…
Grint et al medrxiv.org/content/10.110…
Patone et al medrxiv.org/content/10.110…
Bager et al (Denmark) papers.ssrn.com/sol3/papers.cf…
And others gov.uk/government/pub… 11/16
Challen et al bmj.com/content/372/bm…
Grint et al medrxiv.org/content/10.110…
Patone et al medrxiv.org/content/10.110…
Bager et al (Denmark) papers.ssrn.com/sol3/papers.cf…
And others gov.uk/government/pub… 11/16
What does this mean? First, it helps to explain why the UK has seen so many COVID-19 deaths in the last three months, together with the increased infectiousness of B.1.1.7, as we described here: science.sciencemag.org/content/early/… 13/16
Second, it adds more evidence that other countries may have a difficult time in the months ahead. Vaccines are effective against B.1.1.7 and will help a lot.
science.sciencemag.org/content/371/65…
papers.ssrn.com/sol3/papers.cf… 14/16
science.sciencemag.org/content/371/65…
papers.ssrn.com/sol3/papers.cf… 14/16
Thanks to: coauthors @Jarvis_Stats John Edmunds @NP_Jewell @karlado @RuthHKeogh; @cmmid_lshtm & @LSHTM; peer reviewers; @rjchallen, @leondanon, SPI-M & NERVTAG for discussions. Big thanks to @PHE_uk for allowing us to release anonymised data: github.com/nicholasdavies… 15/16
And for funding support, thanks to @NIHRresearch @UKRI_News @The_MRC @GCRF @ESRC @EU_Commission @NIH @NIAIDNews @royalsociety @wellcometrust. 16/16
• • •
Missing some Tweet in this thread? You can try to
force a refresh