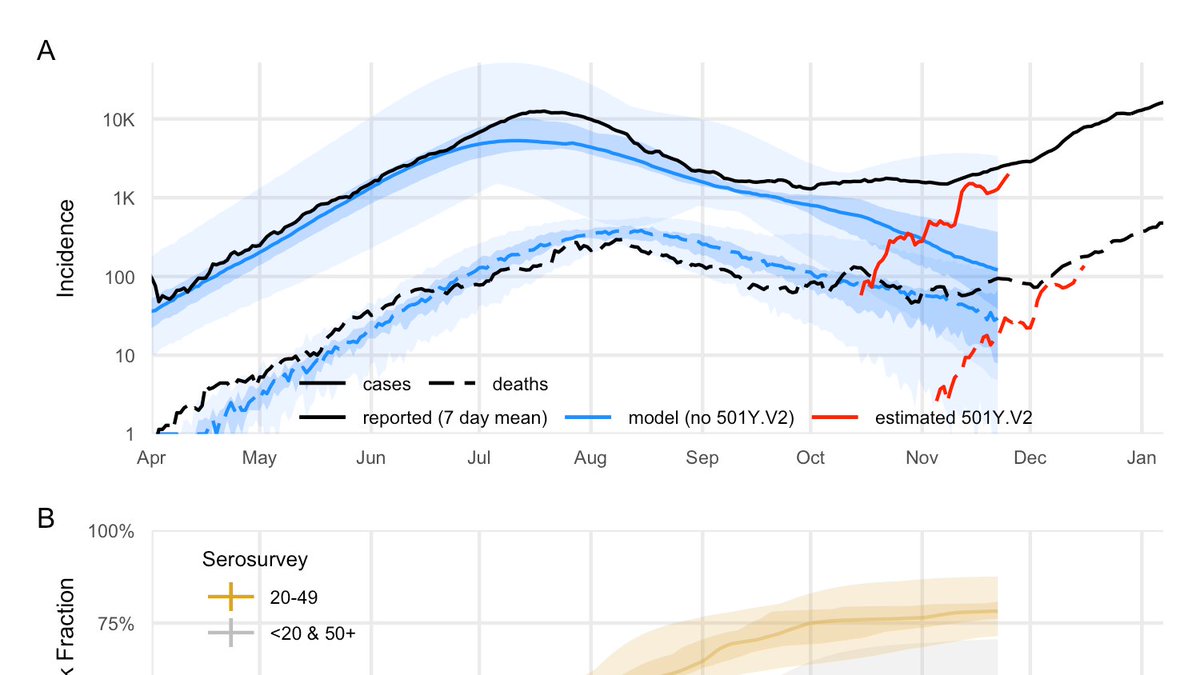
As discussed at #EPIDEMICS8 breakout session, @saCOVID19mc have started characterizing the #SARSCoV2 variant #Omicron; working estimates for this new variant are critical so the globe can plan with the best information possible: drive.google.com/file/d/1hA6Mec…
#NotYetPeerReviewed
#NotYetPeerReviewed
Based on S-Gene Target Failure (SGTF), we estimated #Omicron growth rate across RSA provinces. Gauteng has the clearest signal, and indicates growth of 0.21 (0.14, 0.27)/day, corresponding to a doubling time of 3.3 (2.5, 4.6) days.
#NotYetPeerReviewed
#NotYetPeerReviewed

From that, we estimated relative effective reproductive #s for #Omicron & #Delta. In Gauteng, the ratio is ~2.4 (2.0, 2.7), assuming #Omicron has the same typical time between generations.
[n.b. this corrects fig from pres.; no change to other results]
#NotYetPeerReviewed
[n.b. this corrects fig from pres.; no change to other results]
#NotYetPeerReviewed

Using next generation matrix approx of Gauteng, incorporating accumulated immunity due to vaccination and infection, we can constrain the mix of transmissibility & immune escape for #Omicron, under various plausible scenarios.
#NotYetPeerReviewed
#NotYetPeerReviewed

We hope researchers around the globe will be able to use these estimates to focus their analysis in the coming days & adequately prepare.
This is evolving work as more data comes in, and we will update as we learn more.
#NotYetPeerReviewed
This is evolving work as more data comes in, and we will update as we learn more.
#NotYetPeerReviewed
work with @SheetalSilal, @jd_mathbio, @seabbs, @carivs, @JemBingham, @hivepi, @SACEMAdirector and authors not on twitter; special thanks to @DubzeeLi and @sbfnk.
@threadreaderapp please unroll.
@threadreaderapp please unroll.
• • •
Missing some Tweet in this thread? You can try to
force a refresh