With new analyses showing lower severity for Omicron cases, what will this mean for our projections? Well obviously this will tend to bring the numbers lower, which is great — but I want to clear up a few misconceptions.
https://twitter.com/BarnardResearch/status/1469646833563615237
There was some confusion over what our models, which assumed Omicron had equal "baseline" severity to Delta, meant for severity in practice. This leads to around a 40% reduction in realised severity within each age group, because more Omicron cases are breakthrough/reinfections. 

There was even a @jpmorgan reanalysis of our projections that just cut the hospitalisation numbers in half, because of reports from South Africa that the hospitalisation rate was lower. We were already predicting that the average hospitalisation rate would be lower (see above).
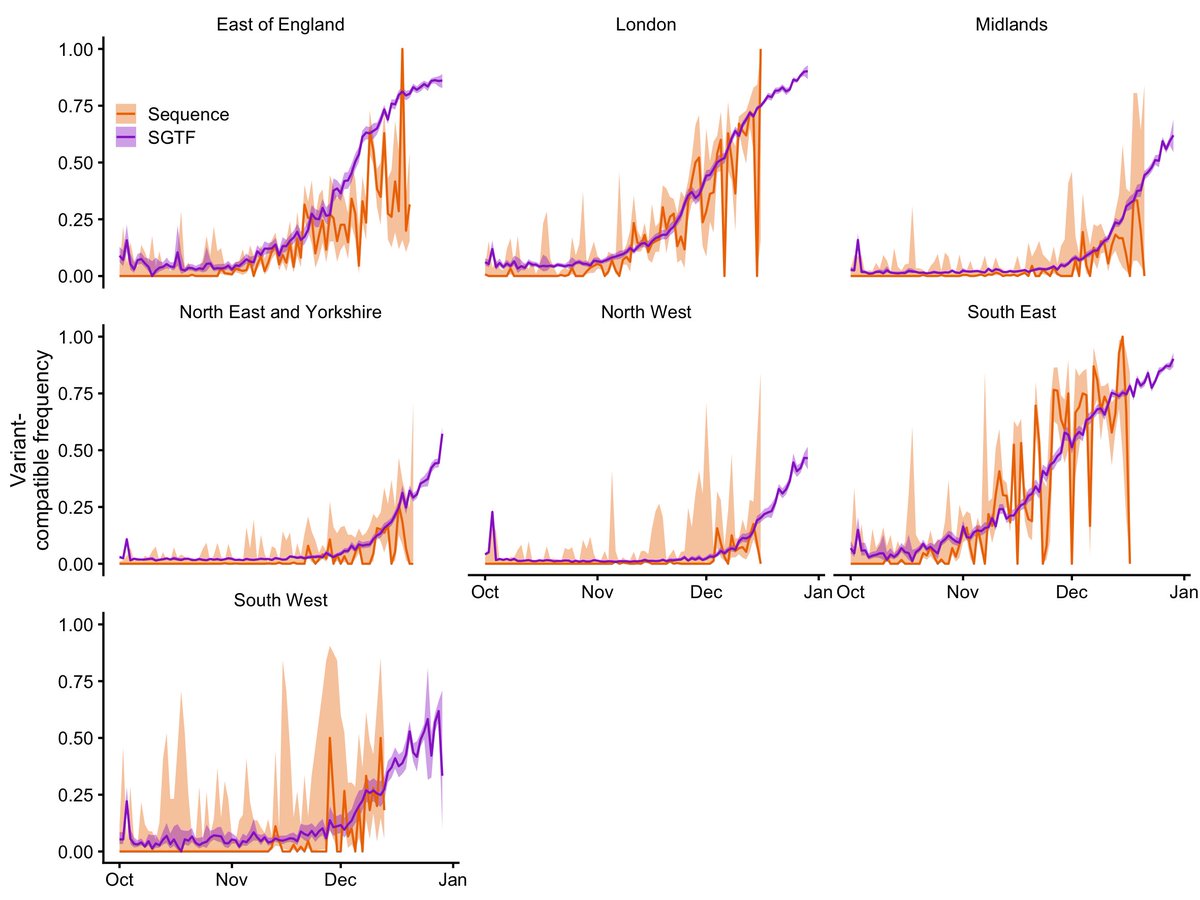
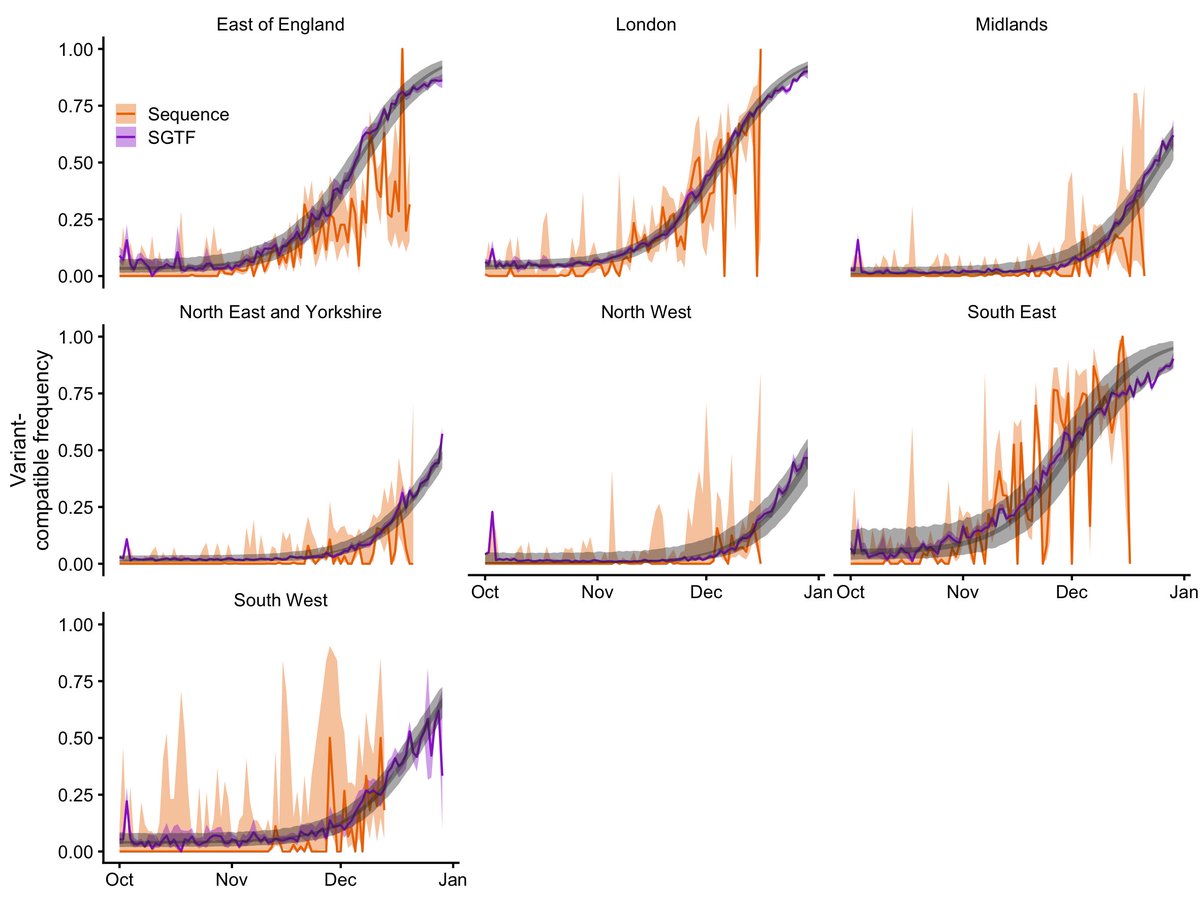
New papers by @imperialcollege (report 50) show that A&E visits among Omicron cases are reduced by 30-40%. The former would be "70% baseline severity" which looks like this in practice (these plots are outputs from our model btw which we will add to the next preprint version) 

There is a really important point, though, that some hugely helpful explainers @JamesWard73 @nataliexdean @jburnmurdoch have so far not really focused on (not saying they have overlooked this, just that I haven't seen them tweet about it yet...). And here it is:
The same principle that makes Omicron relatively less severe WITHIN each age/risk group—its better ability to infect those with preexisting protection—will tend to shift the case mix, relative to Delta, AMONG the age/risk groups into those that have relatively better protection.
For a lot of countries, including the UK, this means that the Omicron wave will tend to infect *relatively* more vulnerable older people than the Delta wave, which after all was really primarily in young people.
To the extent that this shift happens, the overall realised severity across all age groups (i.e. the average IHR) will not decrease as much as the above plots suggest.
If the fraction of highly-protected older people infected with Omicron is big enough relative to the very small fraction who got Delta, then in theory this could even increase the average IHR even though it is decreasing within each age group. en.wikipedia.org/wiki/Simpson%2…
If in South Africa, there was higher protection among young people prior to Omicron, this same effect would tend to *decrease* the IHR resulting from Omicron if it was shifting the average Omicron case younger relative to Delta.
The older and vulnerable age groups drive the majority of morbidity and mortality coming out of any model of SARS-CoV-2 transmission. If people exercise even a little bit of caution around their vulnerable/elderly relatives this Christmas, that will make a HUGE difference.
Hence the importance of testing yourself before seeing your friends and family and before / after going out to places where the risk of transmission is high.
• • •
Missing some Tweet in this thread? You can try to
force a refresh