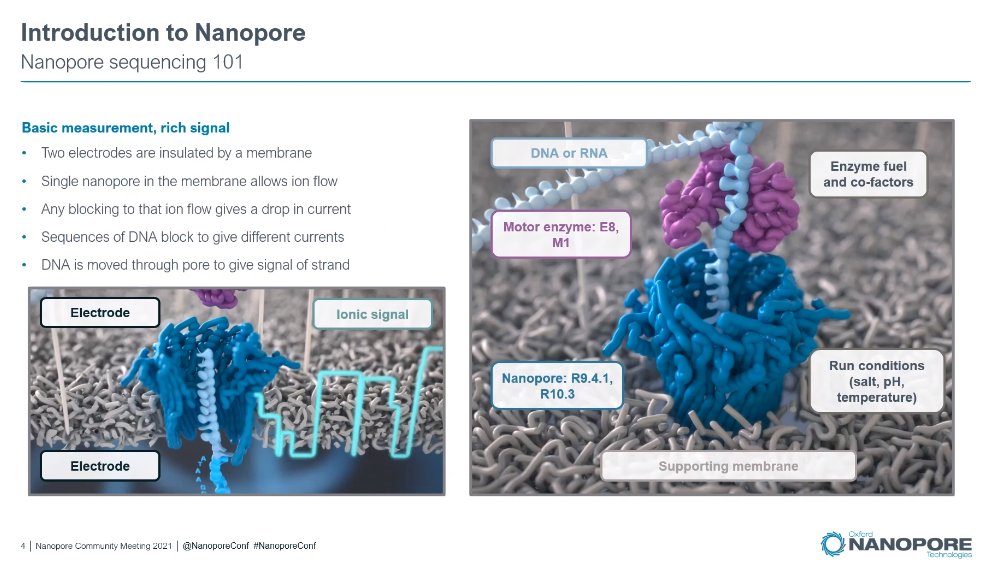
One more from #JPM22, maybe the last one from me: PacBio's presentation which took place on the 13th January. Intro slide about the company. 

There are now 374 Sequel II/IIe machines installed, the company's revenue is growing and is in the triple digit annually. 

The number of SMRT Cells shipped has almost doubled since 2020, quadrupled since 2019. That's a good sign for PacBio. 

The company describes 2021 as the most productive year for the company. Highlights are the acquisition of Omniome's Sequencing by binding (SBB) short-read technology, and partnership with Invitae. 

Important improvements in sample prep and data analysis to make the HiFi technology more attractive. 

Interesting that the partnership with Google Health is not only on DeepVariant calling, but also how the software from Google deep learning improves the callability of reads. Base calling is at the center of the data analysis efforts of #NGS companies. 

Announcement of the "desktop HiFi-based instrument" in partnership with Berry Genomics in China, who want to use it for NIPT and other assays. Someone described this would have the profile/size of a NextSeq instrument. 

PacBio takes a short-read play because they anticipate it will remain relevant in certain field, e.g. cfDNA Liquid Biopsy for early detection of cancer. 

The Omniome's SBB technology is already being pushed beyond 150bp, showing a different clustering method to the original emPCR (nobody likes emulsion PCR). 

Over the read cycles, the mismatch rate remains constant and very low. I don't know how to interpret this compared with the previous slide, other than maybe the errors are uniform in type by Q-scores go down around 200 cycles? @sbarnettARK @OmicsOmicsBlog 

The first SBB instrument is in partnership with Invitae. I think this is also expected to be the size of a NextSeq, and depending on the throughput, yet another copycat (in instrument size) to the list together with Singular Omics G4. 

So lots to go for PacBio, I am quite keen to see how both fronts, the HiFi and SBB technologies, evolve. 

If I compare PacBio's strategy with what Illumina, MGI Tech and Oxford Nanopore are doing, I would say that: it's commendable that they take the short-read approach seriously, as the other 3 companies have a short-read strategy themselves.
If the HiFi technology continues to improve in throughput (number of molecules) and cost, it will continue to have a market out there. But recent data from @AW_NGS in the Netherlands shows that Oxford @nanopore's Q20+ ultralong reads could now be giving better assemblies than ...
... the best PacBio can do. This was in some rather large and repetitive plant genomes, which is where the extra read length will give better value to the assembly process, but I think it extrapolates overall to any type of assembly task. Keen to see more data on both sizes here.
We should keep in mind that human resequencing has been historically done with Illumina due to cost considerations, but this could switch to long read technologies once the difference narrows. "Sequence once keep forever" for human reference genomes could...
... mean in the future we all have our genome sequenced with long reads, best technology solution, and each of our genomes is a de novo assembly. It would be in our electronic health records (we could carry a copy in our mobile phones) and our GP will refer to it during our ...
... lifetime. This would be in comparison with regular epigenomic health checks, probably mostly cfDNA from blood draws, 10-20ml in volume or the size of a lipstick. These will be short fragments and will contain epigenomic marks that relate to our physiological health status.
If they done regularly, they could alert our GP of an early cancer detected in one of our tissues/organs, which will refer us to the oncologist. They could alert us of neurological pathologies or cardiovascular issues that will also need follow-up.
So overall, this long-reads "sequence once, keep forever" initial sequencing of each of us will then generate the reference for the regular epigenomic short-read sequencing, and could become a standard in a few years time.
All in all, companies in the #NGS space should think about having a play in both short-reads and long-reads. Oxford @nanopore has shown they can produce short-reads at a very good tick with the same technology that produces long and ultralong reads.
PacBio seems to want to combine the parallel HiFi + SBB approach, so two technologies under the umbrella of the same company.
Illumina is the king in short-reads, and after the failed attempt to acquire PacBio, has now come up with an Infinity 10Kb reads method, the details of which we hope to learn more soon. If this will be enough and compare well with the other approaches, time will tell.
Finally MGI Tech has a very good short-read technology, in some ways already better than Illumina, and has demonstrated their stLFR technology, which improves the haplotypic information of the short reads. en.mgi-tech.com/products/reage…
The other companies don't seem to be at the same level as these four yet, but there are definitely another 4-5 contenders here that could jump to prominence at any point. Looking forward to see what the future brings.
• • •
Missing some Tweet in this thread? You can try to
force a refresh