"[...] everything apart from the computer is designed by Nanopore, but [the computer side] may change in the future, by the way". Unreasonable to think they will build their own silicon specialized for base-calling?
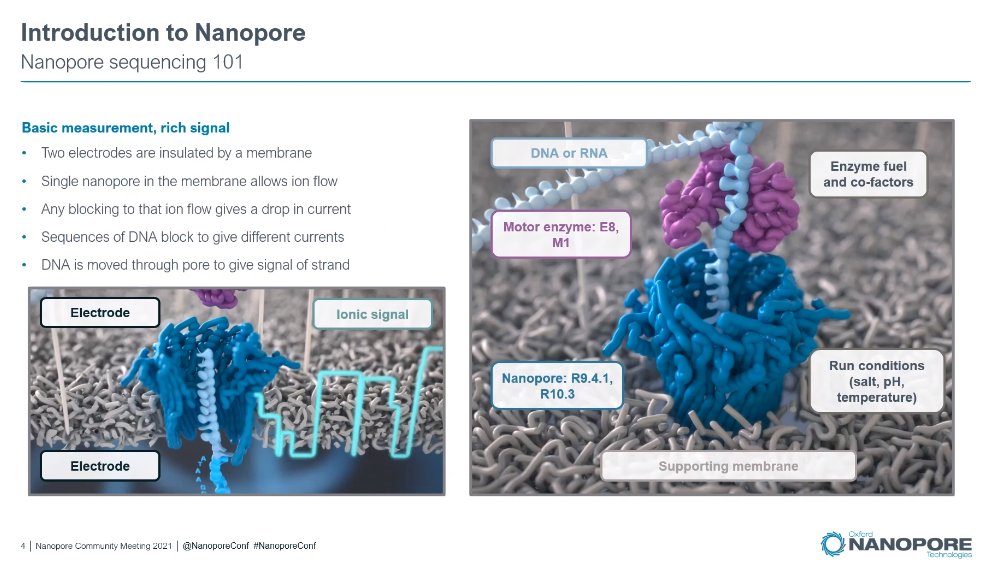
A pet-peeve of Clive, having to explain that #Nanopore technology does not mind the read length, i.e. Read Length == Fragment Length. 

Smallest pore sliding window would be 20-30 bases. Generally speaking the data is the same. Always been capable of doing short fragments.
Short fragment mode is a collection of software tweaks (targeting beginning 2022). More than 250 million native human reads from a single PromethION flowcell. 

Competitive with the market: >20M reads MinION, >160M reads PromethION. My back of the envelope calculations is that for 166bp cfDNA, this would generate about 40Gb of data. 

Accuracy: extracting value from the signal is the key. First time we see the new fish, Remora, mentioned for modified bases (Bonito v0.5 released just now) 

Q20+ is chemistry+software update. Released earlier in 2021, now the combination of R10.4 + E8.1 now achieves a modal of more than Q20. This is called kit12 in the product page. All one strand, one pass (a.k.a. simplex). 

A bit slower, not as quick as the old chemistry. It's been out for a while as Early Access, but not GA in the website shop.
The + in Q20+ largely coming from software improvements, probably more to achieve on that front. But duplex mode, reading from both strands of a dsDNA is at Q30. Consensus-wise, Q50 bacterial genomes at 20x coverage (lower than the previous iteration). 

Chemistry + Training set composition + Latest Machine Learning software tools. New mode "Super Accuracy" on top of the "High Accuracy" mode. 

Speed of Bonito also improved, so smaller distance performance-wise to the production Guppy caller.
Duplex: combining data from both strands. AVailable as Early Access for a few weeks/months. Adapters on both ends. Joined calling of the molecule combining signal from both strands. Internally optimizing efficiency, currently around 50%. 

New kit in January 2022 will push efficiency of duplex at around 40%. Not affected by fragment length. A bit lower in native DNA, possibly due to nicks in dsDNA already present in the sample. 

WIP on duplex software side. More work to do, but now that Simplex caller has been improved, effort goes into Duplex. Guppy (beta feature). A new version of Bonito should soon be out (is it in v0.5?) 

Very high accuracy on very long reads. This is a differentiator from PacBio HiFi, where there is an intrinsic trade-off between multi-pass and length of the insert. 

Mod bases, covered by Markus' webinar. Better than bisulphite, which was the gold standard in the past. 

These extra letters in the alphabet complicate matters in the Bioinformatics tools downstream. Remore optional choice of output file (fastq, sam, bam, cram). In Bonito now both 5mC and 5hmC (quite frankly, everything that matters for human epigenomics). 

*Remora
Improve the pore itself: e.g. longer pore should give better accuracy. Homopolymers being chased down. 

A dial to choose what you need: Accuracy vs Speed. Small impact on accuracy by changing temperature, but gives yield benefits. 

We can phase out the old R.9 and just give a flexible R10.4 or more in terms of accuracy and speed.
Inny vs Outy: exploring the plasticity of what to do better given the same hardware, as these are changes in the chemistry/enzymes. 

Outy sequencing introduction, now working at ~200 bases/second. There is a "wait" state which can be used to make a decision: eject or do something else. 

The threading step is useful to size the strand: e.g. decide what size to do, discard the rest.
But biggest difference is ability to do multi-pass on the same molecule. Very naive software and primitive pore at the moment, but showing a lot of promise. 

Given that this is software-controlled, you have real-time control: basecall -> assess -> basecall -> assess, until you have the desired Q-score, then eject molecule and start with the next one.
Given what we already know from adaptive sampling, we can decide how much to focus on which strands. E.g. oncogenes within a WGS sample, or rare species sequence in river water, etc. 
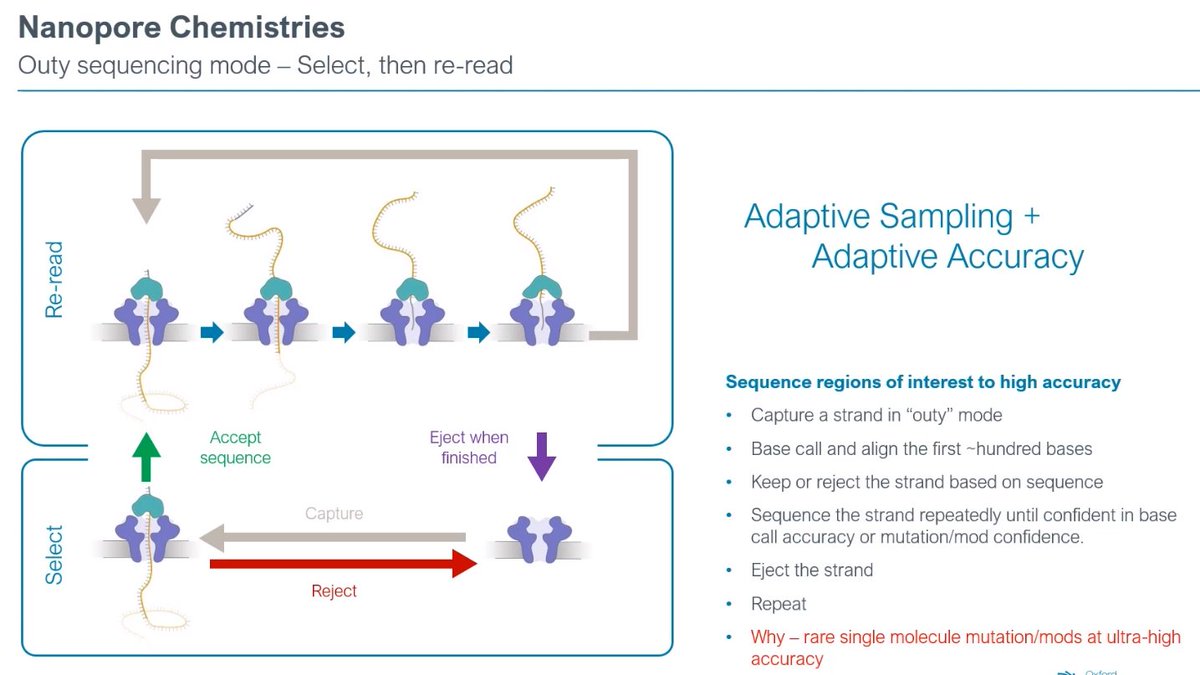
Adaptive sampling + Adaptive accuracy for each application. Needle in the haystack applications. Come to think of it, for low frequency mutations, this is better than anything else...
Aiming at a Liquid Biopsy Platform (device). Extra sample chamber to enable applications with small input amount of DNA. Potentially Point of Care or Point of Field device for #LiquidBiopsy. 

Current prototype. This was mentioned in AGBT 2012. Still a lot of work to do but much happier now in the direction. 

Browser Calling: making use of WASM/WebGPU technologies. Built with TensorFlow.js. Nice little idea to remove the complexity away from installation/deployment. 

All done local, no cloud assistance. No installation faff.
MinION Mk1D. Essentially an iPad Pro connected to a flattened MinION. Large battery life which is useful for field work. Importantly 5G connectivity. WASM Bonito works with M1-Max. 

• • •
Missing some Tweet in this thread? You can try to
force a refresh