Now that @Quantum_Si has given us a peek under the hood of its protein #sequencing platform (Platinum), we can begin comparing actual results to theory.
A few months ago, I shared this paper that gave a theoretical framework for protein sequencing:
pubs.acs.org/doi/10.1021/ac…
A few months ago, I shared this paper that gave a theoretical framework for protein sequencing:
pubs.acs.org/doi/10.1021/ac…
The author simulated how different factors, such as the # of readable amino acids (AAs) and the read length, would affect a protein sequencer's ability to unambiguously detect the 20,000 canonical human proteins in our bodies.
That chart is attached below.
That chart is attached below.
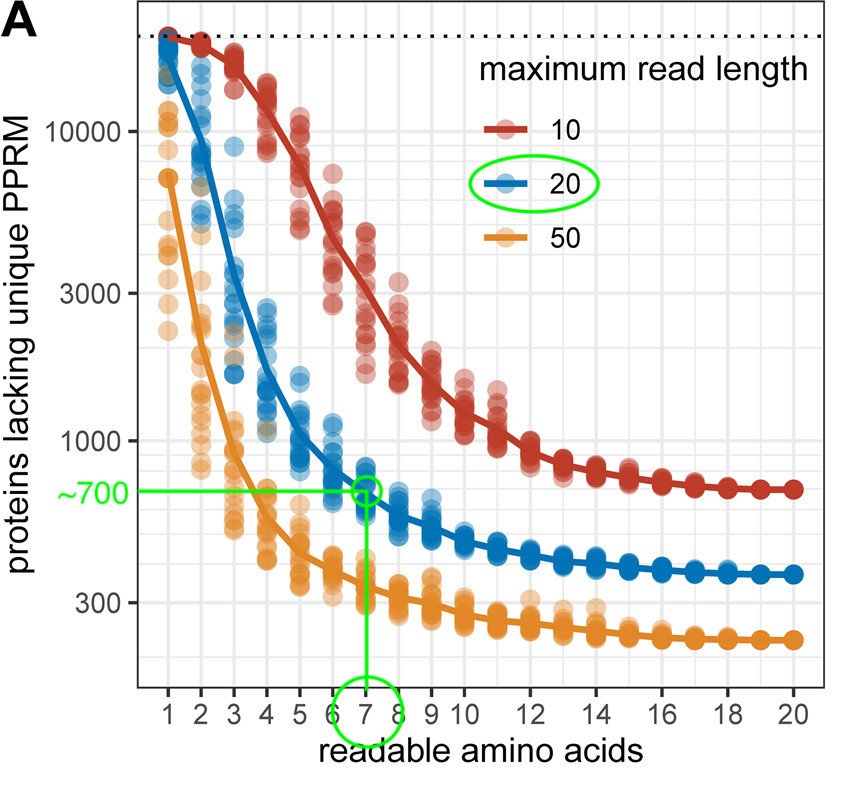
I've marked in green where QSI currently stacks up. Based on its recent pre-print (linked below), Platinum can directly read seven (7) amino acids (F, Y, W, L, I, V, and R) with peptide reads that seem to max out around 20 AAs.
biorxiv.org/content/10.110…
biorxiv.org/content/10.110…
Based on my crude line drawings, QSI may be able to detect 19,300/20,000 proteins (~96% of total) despite only having a repertoire of 7 distinct AA recognizers. Indeed, that's about what they claim in the paper. 

Of course, there are pragmatic concerns that need addressing to truly battle-test this number, namely loading efficiency (Poisson limits), dynamic range compression (Howdy, Seer!), CMOS chip density, etc.
I'll share more on these when early-access data starts accruing.
I'll share more on these when early-access data starts accruing.
It's also interesting to see which seven AAs QSI can detect, which I've marked in the chart below. Some AAs (given their rarity) matter more when discriminating between proteins. QSI's seven seem to span the entire range from not-very to very important! 

I'm eager to see just how much users can get done with the performance as it currently stands. We believe QSIs potential to read substantially all AAs (and post-translational modifications PTMs) is astounding. As such, I plan to track the company's progress ...
...as it engineers more AA recognizers (seems like a good @Ginkgo project) to round out Platinum's protein sequencing capabilities.
For targeted sequencing work, I think the potential at launch is tangible.
I'm murkier on the prospects (in the near term) for protein counting.
For targeted sequencing work, I think the potential at launch is tangible.
I'm murkier on the prospects (in the near term) for protein counting.
As others (@new299) have written, CMOS chips are quite hard to pack densely with sensors (and occupy each one with a single target molecule). As such, the overall throughput may be limited.
41j.com/blog/2021/08/q…
41j.com/blog/2021/08/q…
Even so, a technology that can detect protein variants (proteoforms) at single-molecule sensitivity even in complex mixtures is a tantalizing possibility that I'm excited to watch transition into reality.
• • •
Missing some Tweet in this thread? You can try to
force a refresh