
There’s a lot of magicking and buzzwording around #AI and #MachineLearning in #drugdiscovery
I thought I’d do a thread on a concrete example of using ML to discover a drug candidate about to enter Phase 2a for a stroke disorder:
@RecursionPharma’s $RXRX REC-994
I thought I’d do a thread on a concrete example of using ML to discover a drug candidate about to enter Phase 2a for a stroke disorder:
@RecursionPharma’s $RXRX REC-994
It’s in this Circulation paper from 2015 and illustrates the seed that became $RXRX “phenomics” platform with Chris Gibson (CEO, co-founder) and his former academic mentor Dean Li (board & co-founder) as first/last authors
ahajournals.org/doi/10.1161/ci…
ahajournals.org/doi/10.1161/ci…
The disease:
CCM (cerebral cavernous malformation) is a monogenic loss-of-function stroke disease affecting 1 in 200-500 in the US. 20% are due to the familial form usually due to a loss-of-function mutation in 1 of 3 genes, including the CCM2 gene
CCM (cerebral cavernous malformation) is a monogenic loss-of-function stroke disease affecting 1 in 200-500 in the US. 20% are due to the familial form usually due to a loss-of-function mutation in 1 of 3 genes, including the CCM2 gene
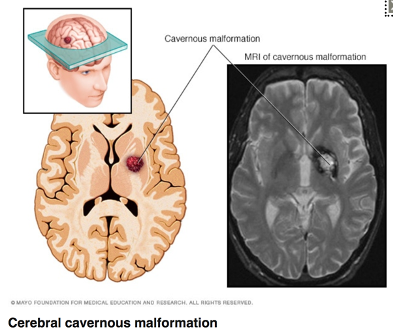
Unstable and leaky vascular malformations in the central nervous system strongly increase the risk of a bleeding (haemorrhagic) stroke (picture).
Other than neurosurgery, there is no treatment for CCM
Other than neurosurgery, there is no treatment for CCM

Previous hypothesis-driven drug discovery approaches had failed. So the researchers tried an ML-based hypothesis-free, unbiased drug repurposing strategy to see if any of a large number of known drugs/molecules might be suitable/repurposed to treat CCM
They plated human blood vessel (microvasc endothelial) cells and used siRNA to knock down the disease-causing CCM2 gene or control siRNA.
So there are “diseased” cells that mimic CCM pathology (right) w a distinct structure and functional phenotype, and “healthy” cells (left)
So there are “diseased” cells that mimic CCM pathology (right) w a distinct structure and functional phenotype, and “healthy” cells (left)

All cells were stained with a few standard dyes for nucleus, membrane, actin &c and an automated microscope was used to take a large number of fluorescent microcopy pictures.
So there are many images of diseased cells, and many of healthy cells
So there are many images of diseased cells, and many of healthy cells
They used the open-source software CellProfiler (by @DrAnneCarpenter et al at the Broad) to automatically identify the borders of cells and create a database of a large number of mathematical descriptors for every cell in every image
Such descriptors could be anything derivable from the stained cells images.
For example, the radial distribution of a stained adhesion molecular called VE cadherin (right), or the number of neighbours for each cell, or % cell borders touching other cells (left)
For example, the radial distribution of a stained adhesion molecular called VE cadherin (right), or the number of neighbours for each cell, or % cell borders touching other cells (left)

The researchers then used an ML tool called CellProfiler Analyst to “learn” a set of rules to distinguish diseased (CCM2 knockdown) from healthy cells. It’s a type of *supervised* ML, where the algo knows the outcome (healthy/diseased) for each cell in the training set.
The researchers then exposed diseased (ie CCM2 knockdown) cells to one of 2,100 known drugs and bioactive compounds and acquired high-throughput fluorescent microcopy images for each
The images where then fed to the ML algo asking “do they look like healthy cells” to identify compounds which had “rescued” the disease phenotpye. ML did that by applying the rules it previously learned classifying each image by the % of cells scored as CCM2 knockdown or control
They selected the top 38 compounds identified by ML as rescuing the disease phenotype
(They also had 2 humans eyeball images and choose compounds, but they didn’t do so well at picking successful candidates)
(They also had 2 humans eyeball images and choose compounds, but they didn’t do so well at picking successful candidates)
They then tested the 38 compounds using a more traditional, low-throughput transcellular resistance assay (CCM makes cells “leaky”) that narrowed the top candidate list down to 7 

Next, they injected the 7 compounds as transdermal wheals into endothelium-specific Ccm2 knockout mice and found that 2 reduced peri-injection site microvascular leakiness 

Finally, they tested these 2 compounds in a more elaborate chronic animal model of CCM using the same type of Ccm2 knockout mice.
Both top compounds (tempol/REC-994 and Vitamin D3) reduced cerebrovascular lesions on MRI
Both top compounds (tempol/REC-994 and Vitamin D3) reduced cerebrovascular lesions on MRI

REC-994 (tempol) is a superoxide scavenger which $RXRX are about (1Q22e) to take into phase 2 clinicaltrials.gov/ct2/show/NCT05…
This ~10yo example demonstrates how a high-throughput, simple, cheap ML-based primary screen of microscopy images can be used to comb through
This ~10yo example demonstrates how a high-throughput, simple, cheap ML-based primary screen of microscopy images can be used to comb through

a huge number of compounds that could never reasonably be tested using traditional, low-throughput assays.
It also shows how a hypothesis-free approach can identify unexpected targets – tempol/REC-994 would never have been chosen based on its previously known biology
It also shows how a hypothesis-free approach can identify unexpected targets – tempol/REC-994 would never have been chosen based on its previously known biology
The foundation of $RXRX current approach to AI/ML drug discovery is still the automated acquisition of a huge number of microcopy images from cells w CRISPR/Cas9-knocked genes, treated w compounds &c – supplemented w -omics data and done at enormous scale 

• • •
Missing some Tweet in this thread? You can try to
force a refresh





