Element Bio (@elembio) is announcing their first instrument today. I'll concentrate on my own highlights on the presentation, which I am sure it'll be available as a video later on. 

My professional interest on the announcement today is: could this be a replacement to a mid-throughput instrument like the NextSeq 2000/1000? If the price in the first specs slide is right, I think the answer is yes! 

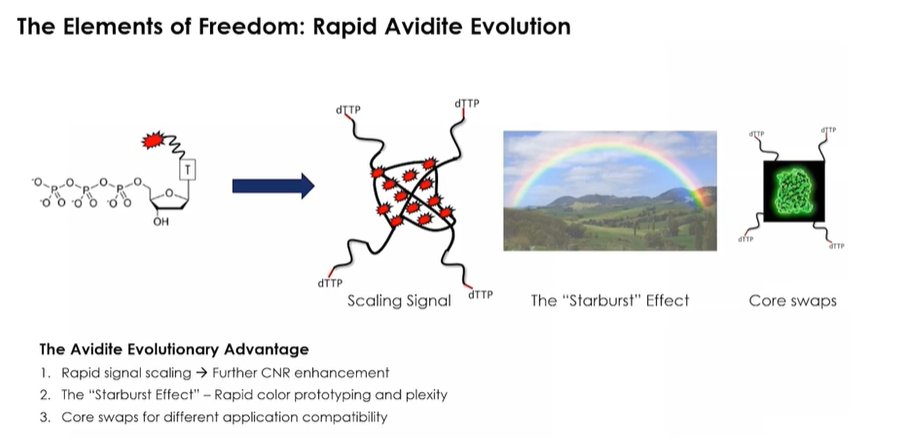
Surface chemistry is low binding, which means the proteins from previous cycles don't attach to it, lower background. Description of the Avidity Advantage. 

A difference of 100-fold reduction in reagent consumption per cycle, which makes the system more cost effective than "traditional methods". The multivalency is key: the fluorophore is attached to many avidites. 



On the optics, they managed to use better optical properties from the new CNR. Data quality impressive. 





Sample prep and analysis. On the former, we've already seen a plethora of announcements with other companies. Primary analysis gives FASTQ files ready for the next step. Multiplexing to up to 96plex, minimal index hopping. 

Very cleverly, there is a library conversion process: take existing (Illumina) libraries and convert them so they can be sequenced on the Element Bio AVITI instrument. 

Tunable output: I think this means you can interrupt a run if it's not going well, and reuse the remaining reagents later on. 

Element Bio announced recently that they acquired Loop Genomics, a SanFran company that creates synthetic long read molecules from short-read readouts. Importantly, they will continue selling to non-Element Bio customers. 

The sample prep kit has all needed for taking the long molecules into an NGS sequencer, then analyse via the associated software to transform into the synthetic long reads. 

Loop barcode with a UMI attaches to the long molecule, it is then amplified, and then the "Distribute" step, which is a copy+paste step of the barcode "within" the long read molecule. 

Each cluster of short reads with the same barcode is then assembled in silico to produce long read FASTQ files.
I am thinking how this differs to the Moleculo approach: it's probably bead-based instead of diluting plates, but other than that, the assembly process seems similar. Compared to Longas (Illumina Infinity?) there is no introduced long PCR mutation to "label" short-reads.
LoopSeq Solo: Sanger-like sequencing using NGS. Multiplex your sequences of interest in plates, then put on an NGS instrument, then reconstruct the Sanger-like longer reads. 

The Loop kits with the Avidity kits will allow people to run not only short-read but also synthetic long-reads with the AVITI instrument.
More emphasis on the new surface chemistry with a 10x better CNR. The whole method took only 4 years to develop and implement into the AVITI instrument.
Who is getting AVITI Instruments? First come first served. Wanting to incentivise the first wave of customers to people that show the power of the platform.
3 beta customers that they could disclose, then another round currently ongoing.
Entering the market with a very robust ecosystem.
Christopher Mason @mason_lab on the testing of the AVITI instrument. 

• • •
Missing some Tweet in this thread? You can try to
force a refresh