
Drug discovery for complex diseases like #Parkinsons (#PD) is challenging - we need screenable cellular phenotypes to move faster. Today in @NatureComms, we present an #AI-driven phenotyping platform that identifies #PD hallmarks in patient cells: nature.com/articles/s4146… 🧵(1/11) 

By combining our cell culture #automation, #CellPainting, high-content imaging, and #DeepLearning methods, we built a robust platform for phenotypic profiling and collaborated with @GoogleAI to screen fibroblasts from 91 #Parkinsons patients and matched healthy controls (2/11) 

We profiled images of nearly 6 million fibroblasts, creating the largest publicly available #CellPainting dataset to date at 48 terabytes (3/11)
Then, we applied #ImageRecognition to these millions of images — the same #AI technology that allows a computer to distinguish an image of a cat and a dog. We trained #MachineLearning models to detect morphological phenotypes that separate #Parkinsons vs. healthy cells (4/11)
To our surprise, our platform successfully identified the exact donor that a given sample came from, across different batches and plate layouts — even more sensitivity than we’d imagined! (5/11)
But more importantly, our platform successfully distinguished between images of #Parkinsons cells vs. healthy controls — both LRRK2 (genetic) and sporadic (non-genetic) PD — with an ROC AUC of 0.79+/-0.08 (6/11) 
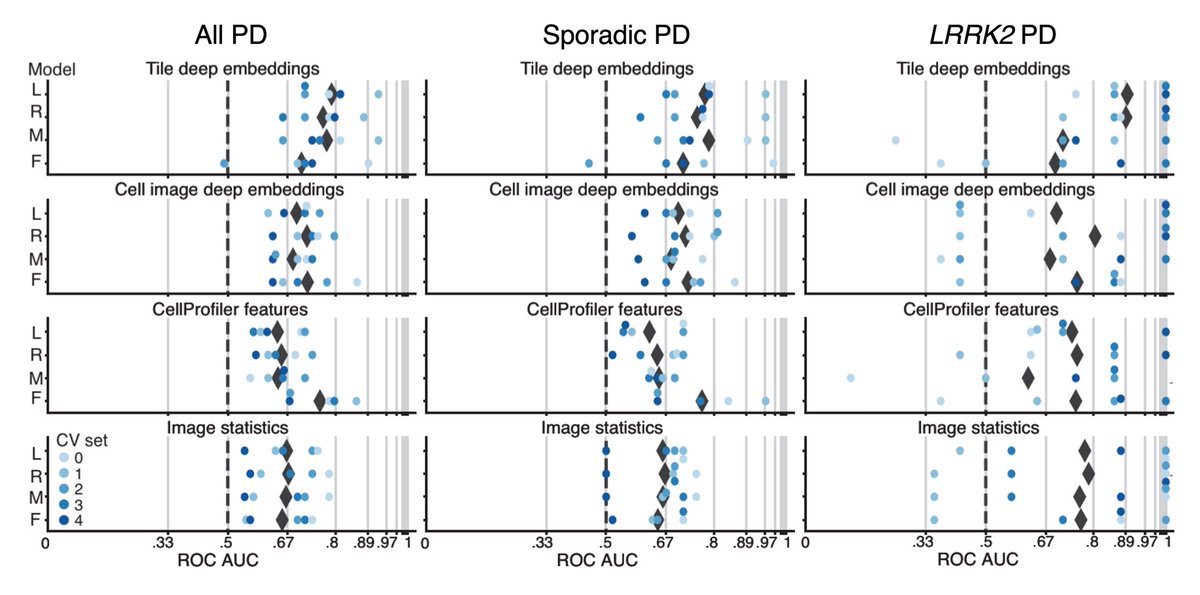
We then asked ourselves: which features drive the separation between patient vs. control? A single organelle? A type of morphology? Turns out it's neither: the features behind these differences are many and subtle, underscoring the complexity of #Parkinsons phenotypes (7/11)
With this agnostic, #AI-driven approach to cellular phenotyping, we have uncovered entirely new features of #Parkinsons otherwise invisible to the human eye (8/11)
Now we're really excited about screening drugs on these cells to reverse these new #Parkinsons phenotypes. And #PD is just the beginning! Since our platform is disease- and cell type-agnostic, we can apply it to #iPSC derivatives, #RareDiseases, & other complex diseases! (9/11)
Huge congratulations to the amazing NYSCF team — biologists, hardware/software engineers, data scientists — and our collaborators @GoogleAI. This is the power of #teamscience! Many thanks to all of our study participants, and to all of you who support NYSCF’s research (10/11)
Finally, we’ve dedicated this paper to the memory of our dear friend and colleague, Reid Otto (1969–2022), an exceptional engineer who helped make this study possible, and exemplified team science and collaboration every day ❤️ (11/11) 

• • •
Missing some Tweet in this thread? You can try to
force a refresh



