#拡散希望 速報🔔 G2P-Japan🇯🇵の最新作「オミクロン亜株XBB.1.5の性状解析」を、論文/プレプリント公開前に速報します(図はメイキング)。本研究では、流行拡大中の#オミクロン XBB.1.5株(通称 #クラーケン)の性質を解明しました🔥 1/
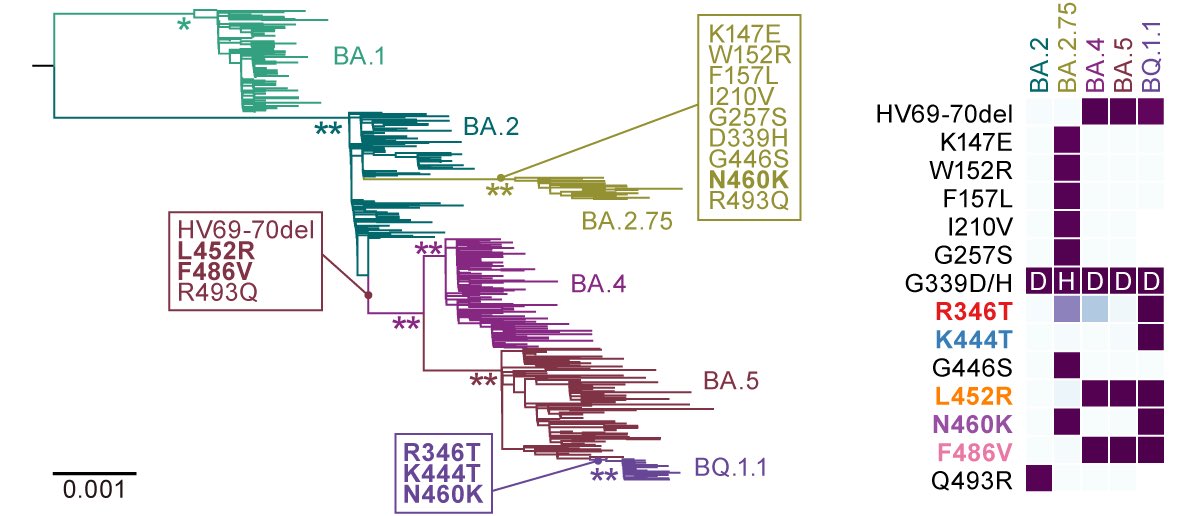
2022年末にアメリカ🇺🇸で突如感染爆発を始めたのがXBB.1.5株です。この株は、↓で我々が報告しているXBB.1株(G2P-Japan🇯🇵プレプリント第15弾)が、スパイクに #F486P という変異を獲得したものです。2/
https://twitter.com/SystemsVirology/status/1607886362807791616?s=20&t=DBwBW_H3J3ZYlTBbPqmMXw
それに加え、XBB系統が持っていた #Y144del という欠損が復帰したXBB.1.5株(XBB.1.5+ins144Y)も確認されています。Y144del という変異は、ブレイクスルー血清への抵抗性の上昇と、ウイルスの感染力の減少に作用する変異です(G2P-Japan🇯🇵プレプリント第15弾より)。3/ 


これは、BA.1とBA.2、BA.2とBA.5の間の差に匹敵する差であり、これからXBB.1.5株が世界で流行拡大していくことが強く示唆されます。なお、ins144Y変異を追加で獲得することによる実効再生産数の上昇は見られませんでした。5/
次に、#F486P と #ins144Y がスパイクのACE2結合力とシュードウイルスの感染力に与える影響を評価しました。#F486P は、ACE2結合力を爆上げし(結合係数KDを爆下げし)、シュードウイルスの感染力も3倍以上上昇させました。6/ 



一方、#ins144Y は、それ単独ではシュードウイルスの感染力を約2倍上昇させました(XBB.1 < XBB.1+ins144Y)。しかし、#F486P との相加効果は確認されませんでした(XBB.1.5 = XBB.1.5+ins144Y)。7/
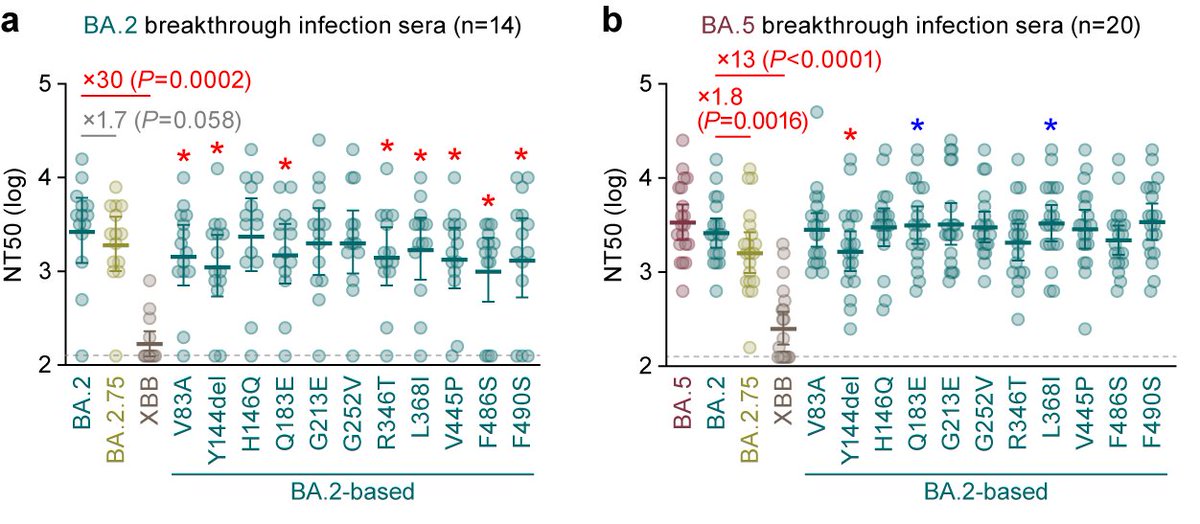
最後に、BA.2, BA.5ブレイクスルー血清を用いた中和試験の結果、XBB.1.5株はXBB.1株と同様、それぞれのブレイクスルー血清に対してきわめて高い抵抗性を示しました。一方、#ins144Y によって、中和抗体抵抗性は減弱しました。8/ 

補足です。
XBB.1.5株のBA.2ブレイクスルー血清に対する抵抗性は、従来株(B.1.1)の41倍、BA.2の20倍で、13人中8人の血清がXBB.1.5に対する中和活性なし。
XBB.1.5株のBA.5ブレイクスルー血清に対する抵抗性は、従来株(B.1.1)の32倍、BA.2の9.5倍で、20人中12人の血清が中和活性なし。9/
XBB.1.5株のBA.2ブレイクスルー血清に対する抵抗性は、従来株(B.1.1)の41倍、BA.2の20倍で、13人中8人の血清がXBB.1.5に対する中和活性なし。
XBB.1.5株のBA.5ブレイクスルー血清に対する抵抗性は、従来株(B.1.1)の32倍、BA.2の9.5倍で、20人中12人の血清が中和活性なし。9/
以上の結果より、XBB.1.5株は、ワクチン血清やブレイクスルー血清による中和作用がすでにほぼ効かないXBB.1株が、#F486P という変異を獲得することにより、ACE2結合力を爆上げしたことによって伝播力を爆上げした変異株だと言えます🔥 10/10
• • •
Missing some Tweet in this thread? You can try to
force a refresh