BREAKING🔔 The 20th paper from G2P-Japan🇯🇵 is out at Nature Communications @NatureComms. We illuminated the convergent evolution of #Omicron and characteristics of BQ.1.1 (aka #Cerberos). Please RT
20 papers in 28 months since the launch of G2P-Japan🎉 1/
nature.com/articles/s4146…
20 papers in 28 months since the launch of G2P-Japan🎉 1/
nature.com/articles/s4146…
In late 2022, the Omicron subvariants have highly diversified. However, some lineages have convergently acquired amino acid substitutions at five critical residues in the spike protein: R346, K444, L452, N460 and F486. 2/
https://twitter.com/dfocosi/status/1575836076089823232?s=20&t=P4COtUsJ2_LztpucW6DS5w
Omicron BQ.1.1 (aka #Cerberus), a descendant of BA.5, bears all five substitutions at the convergent sites: R346T, K444T, L452R, N460K, F486V. 3/ 

Here, we, particularly two young shining stars, @jampei2 & @SpyrosLytras, performed phylogenetic and epidemic dynamics analyses and showed that Omicron subvariants independently increased their viral fitness by acquiring the convergent substitutions. 4/ 

All five convergent substitutions (R346T, K444T, L452R, N460K & F486V) contributed to increase relative effective reproduction number (Re), and BQ.1.1, which harbors all five convergent substitutions, showed the highest fitness among the viruses investigated. 5/ 

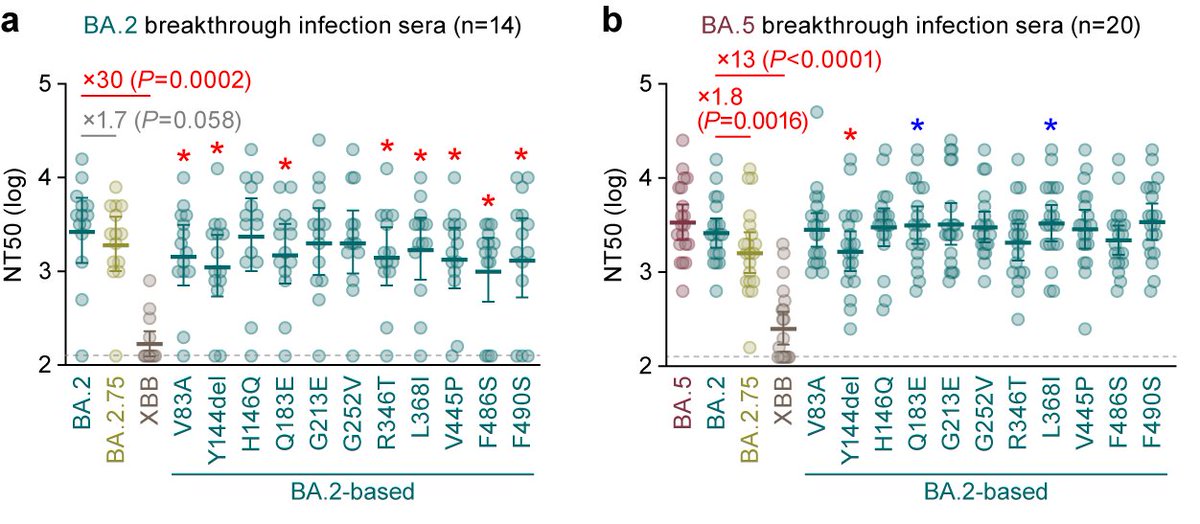
Neutralization assay showed that BQ.1.1 is more resistant to breakthrough BA.2 and BA.5 infection sera than BA.5. 6/ 

We showed lines of data suggesting the close association of viral fusogenicity and intrinsic pathogenicity. For instance, both BA.5 and BA.2.75, descendants of BA.2, are more fusogenic and pathogenic than BA.2. 7/
https://twitter.com/SystemsVirology/status/1569995966954233856?s=20&t=YcTm4-TEfIdF9vaST71ihw
Notably, the BQ.1.1 spike exhibited greater fusogenicity than the BA.5 spike, suggesting that BQ.1.1 is more pathogenic than BA.5. 8/ 
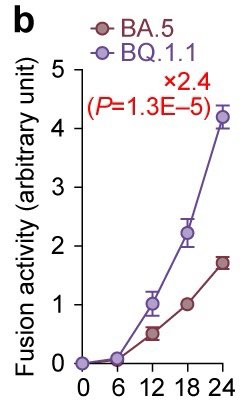
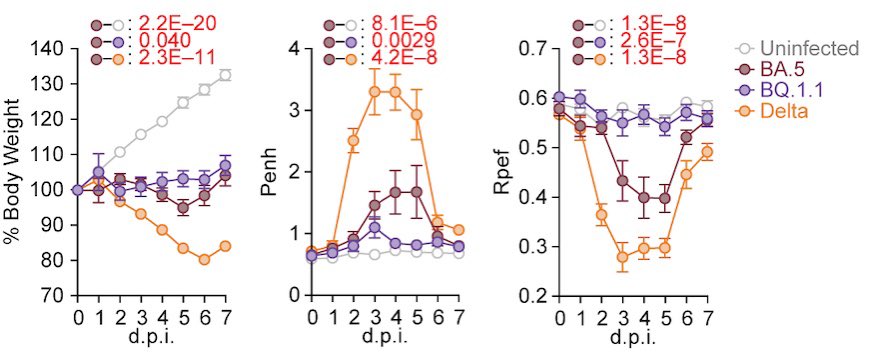
However, unexpectedly, experiments using hamsters showed that the pathogenicity of BQ.1.1 in hamsters is comparable to or even lower than that of BA.5.
Our multiscale investigations provide insights into the evolutionary trajectory of Omicron subvariants. 9/9
Our multiscale investigations provide insights into the evolutionary trajectory of Omicron subvariants. 9/9
• • •
Missing some Tweet in this thread? You can try to
force a refresh

 Read on Twitter
Read on Twitter