
How to get URL link on X (Twitter) App


 Ethylene is a gaseous #phytohormone with a wide range of roles from plant development to immunity. Ernest Starling in 1905 defined a hormone as mobile chemical messenger synthesized by a multicellular organism, that has physiological activity distant from the site of synthesis.
Ethylene is a gaseous #phytohormone with a wide range of roles from plant development to immunity. Ernest Starling in 1905 defined a hormone as mobile chemical messenger synthesized by a multicellular organism, that has physiological activity distant from the site of synthesis. 

 Powdery mildew is a fungal disease of many crop plants, most prominently maybe barley and wheat, where outbreaks can reduce grain quality & yield, and ruin complete harvests. Visible are the fluffy patches formed by the fungus (Blumeria graminis f. sp. hordei).
Powdery mildew is a fungal disease of many crop plants, most prominently maybe barley and wheat, where outbreaks can reduce grain quality & yield, and ruin complete harvests. Visible are the fluffy patches formed by the fungus (Blumeria graminis f. sp. hordei). 

 In the early 1980s scientists finally adopted A. thaliana as model plant. At this point, several mutants were available, but their positions in the genome were mostly unknown. This was years before genome sequences became available,&genetic maps were still based on recombination.
In the early 1980s scientists finally adopted A. thaliana as model plant. At this point, several mutants were available, but their positions in the genome were mostly unknown. This was years before genome sequences became available,&genetic maps were still based on recombination.

 The development of plant transformation in the early 1980s (classics #6&13) was inspirational for many scientists. Among them was Richard Walden, who teamed up with plant transformation pioneers Barbara & Thomas Hohn to leverage this advance to develop the “Agroinfection" method.
The development of plant transformation in the early 1980s (classics #6&13) was inspirational for many scientists. Among them was Richard Walden, who teamed up with plant transformation pioneers Barbara & Thomas Hohn to leverage this advance to develop the “Agroinfection" method. 

 Mendel had always been interested in nature, and grew/kept and observed plants and bees in his parent’s garden. He later decided to become a monk and teacher. However, he failed teacher’s exam in 1850 & 1856, & eventually settled on being a monk and substitute teacher.
Mendel had always been interested in nature, and grew/kept and observed plants and bees in his parent’s garden. He later decided to become a monk and teacher. However, he failed teacher’s exam in 1850 & 1856, & eventually settled on being a monk and substitute teacher.

 Daisy Roulland-Dussoix worked with Werner Arber to study the mechanism for the observed host-specificity of λ Phages. It was known from an important 1953 paper (Bertani & Weigle) that phages, that had replicated in a certain E. coli strain, could only re-infect the same strain.
Daisy Roulland-Dussoix worked with Werner Arber to study the mechanism for the observed host-specificity of λ Phages. It was known from an important 1953 paper (Bertani & Weigle) that phages, that had replicated in a certain E. coli strain, could only re-infect the same strain. 

 I have covered the plant transformation backstory in Classic#6, the T-DNA, so here I will focus on the events after 1983, the year plant transformation was established. These first transformants all were plants regenerated from cultured cells as calli.
I have covered the plant transformation backstory in Classic#6, the T-DNA, so here I will focus on the events after 1983, the year plant transformation was established. These first transformants all were plants regenerated from cultured cells as calli. https://twitter.com/somssichm/status/1447480992176898051
https://twitter.com/somssichm/status/1400315262906159105

 Plants continue to grow for their entire life due to the activity of stable pools of stem cells. The shoot apical #meristem (SAM) is the stem cell niche responsible for producing all above ground cells and is located at the tip of the plant’s stem.
Plants continue to grow for their entire life due to the activity of stable pools of stem cells. The shoot apical #meristem (SAM) is the stem cell niche responsible for producing all above ground cells and is located at the tip of the plant’s stem. 

 In the 1980s it was possible to express transgenes in cells of different organisms, but visualizing & assaying gene expression was still problematic. The lacZ β-galactosidase was a commonly used reporter, but compromised due to endogenous enzymes breaking down its substrates too.
In the 1980s it was possible to express transgenes in cells of different organisms, but visualizing & assaying gene expression was still problematic. The lacZ β-galactosidase was a commonly used reporter, but compromised due to endogenous enzymes breaking down its substrates too.

 Starting with Charles Darwin & son Francis in 1880, several scientists (J. Sachs, F. Went, N. Cholodny...) had speculated that there must be a mobile substance in plants, acting as messenger to direct growth in response to stimuli such as light or gravity (a ‘growth substance’).
Starting with Charles Darwin & son Francis in 1880, several scientists (J. Sachs, F. Went, N. Cholodny...) had speculated that there must be a mobile substance in plants, acting as messenger to direct growth in response to stimuli such as light or gravity (a ‘growth substance’).

 The early 1980s were an important time for #PlantMolecularBiology: Among other things, plant transformation had just been established. But when introducing a gene into a plant, it requires regulatory sequences to activate its expression – and none active in plants were known.
The early 1980s were an important time for #PlantMolecularBiology: Among other things, plant transformation had just been established. But when introducing a gene into a plant, it requires regulatory sequences to activate its expression – and none active in plants were known.

 Jan van Helmont was one of the first scientists who found that the mass of a plant is not acquired from the soil it grows in. When he grew a 5 lb willow tree in 200 lb of soil for 4 years, he found that the tree gained 164 lb, while the soil was only reduced by 2 lb.
Jan van Helmont was one of the first scientists who found that the mass of a plant is not acquired from the soil it grows in. When he grew a 5 lb willow tree in 200 lb of soil for 4 years, he found that the tree gained 164 lb, while the soil was only reduced by 2 lb. 

 In the 1980s, with plant molecular biology still in its infancy, the plant immune system was not understood very well at all. Dangl, at the time an immunologist working on mouse/human cells, remembers: ‘I had never considered that plants could recognize pathogens’.
In the 1980s, with plant molecular biology still in its infancy, the plant immune system was not understood very well at all. Dangl, at the time an immunologist working on mouse/human cells, remembers: ‘I had never considered that plants could recognize pathogens’.

 It was known since before the 1940s, that Agrobacterium could induce tumors (‘crown galls’) on plants, & that these tumors then grow autonomously of the bacterium, meaning that the plant had been permanently ‘transformed’. But the molecular details for the process were not known.
It was known since before the 1940s, that Agrobacterium could induce tumors (‘crown galls’) on plants, & that these tumors then grow autonomously of the bacterium, meaning that the plant had been permanently ‘transformed’. But the molecular details for the process were not known. 

 McClintock started her career & work on #Maize in the 1920s @Cornell, where she immediately made an impact by optimizing chromosome staining methods to characterize the chromosomes of triploid maize in 1929, & then describing the physical basis for chromosomal crossover in 1931.
McClintock started her career & work on #Maize in the 1920s @Cornell, where she immediately made an impact by optimizing chromosome staining methods to characterize the chromosomes of triploid maize in 1929, & then describing the physical basis for chromosomal crossover in 1931. 

 Laibach started work on A. thaliana in 1907, when, for his PhD-thesis, he determined the number of chromosomes in different plants he collected around his hometown Limburg, or @UniBonn, where he worked. A. thaliana only had 5 chromosomes, one of the fewest he found.
Laibach started work on A. thaliana in 1907, when, for his PhD-thesis, he determined the number of chromosomes in different plants he collected around his hometown Limburg, or @UniBonn, where he worked. A. thaliana only had 5 chromosomes, one of the fewest he found.

 Already in 1999/2000, three papers from the legendary Boller-lab @UniBasel in @ThePlantJournal/@MolecularCell identified the elicitor flg22 & its receptor FLS2,laying the groundwork to establish Arabidopsis as a model system to study plant pathogen-interaction & immune-signaling.
Already in 1999/2000, three papers from the legendary Boller-lab @UniBasel in @ThePlantJournal/@MolecularCell identified the elicitor flg22 & its receptor FLS2,laying the groundwork to establish Arabidopsis as a model system to study plant pathogen-interaction & immune-signaling. 

 Mutagenesis is now an invaluable tool to understand a gene’s function. In the early 1900s, when the hereditary substance was not even identified yet, it was an even more important tool, which Emmy Stein added to the small toolbox available to biologists at the time.
Mutagenesis is now an invaluable tool to understand a gene’s function. In the early 1900s, when the hereditary substance was not even identified yet, it was an even more important tool, which Emmy Stein added to the small toolbox available to biologists at the time.

 A regular Arabidopsis flower is composed of 4 whorls, each featuring specific organs: 4 sepals in the outer whorl, followed by 4 petals, then 6 stamen & 2 carpels in the inner whorl. These identities are controlled by the APETALA2 (AP2), PISTILLATA (PI) & AGAMOUS (AG) genes.
A regular Arabidopsis flower is composed of 4 whorls, each featuring specific organs: 4 sepals in the outer whorl, followed by 4 petals, then 6 stamen & 2 carpels in the inner whorl. These identities are controlled by the APETALA2 (AP2), PISTILLATA (PI) & AGAMOUS (AG) genes. 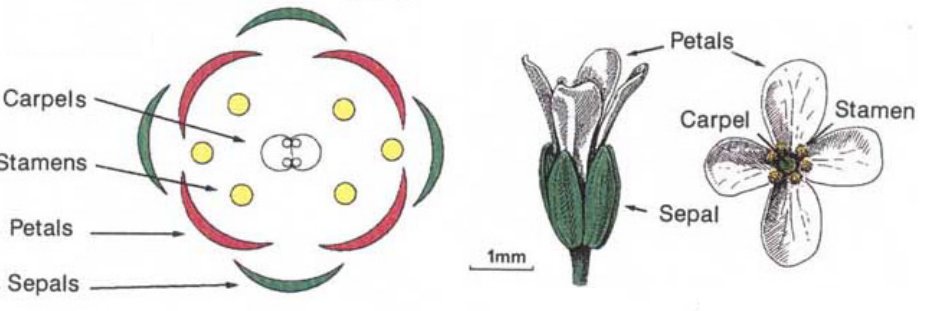

https://twitter.com/somssichm/status/1386558825088552962
 2/14 In 1962 Osamu Shimomura et al. identified the bioluminescent Aequorin in the Aequorea jellyfish, as well as a green fluorescent protein, that seemed to act as a FRET-acceptor for the Aequorin 'in jelly' [1][2][3] (REFs at the end).
2/14 In 1962 Osamu Shimomura et al. identified the bioluminescent Aequorin in the Aequorea jellyfish, as well as a green fluorescent protein, that seemed to act as a FRET-acceptor for the Aequorin 'in jelly' [1][2][3] (REFs at the end). 