
Pathogen entry into cells, host immune responses & vaccine design: structural biology, protein design, virology & immunology @HHMINEWS @UWBiochemistry
How to get URL link on X (Twitter) App


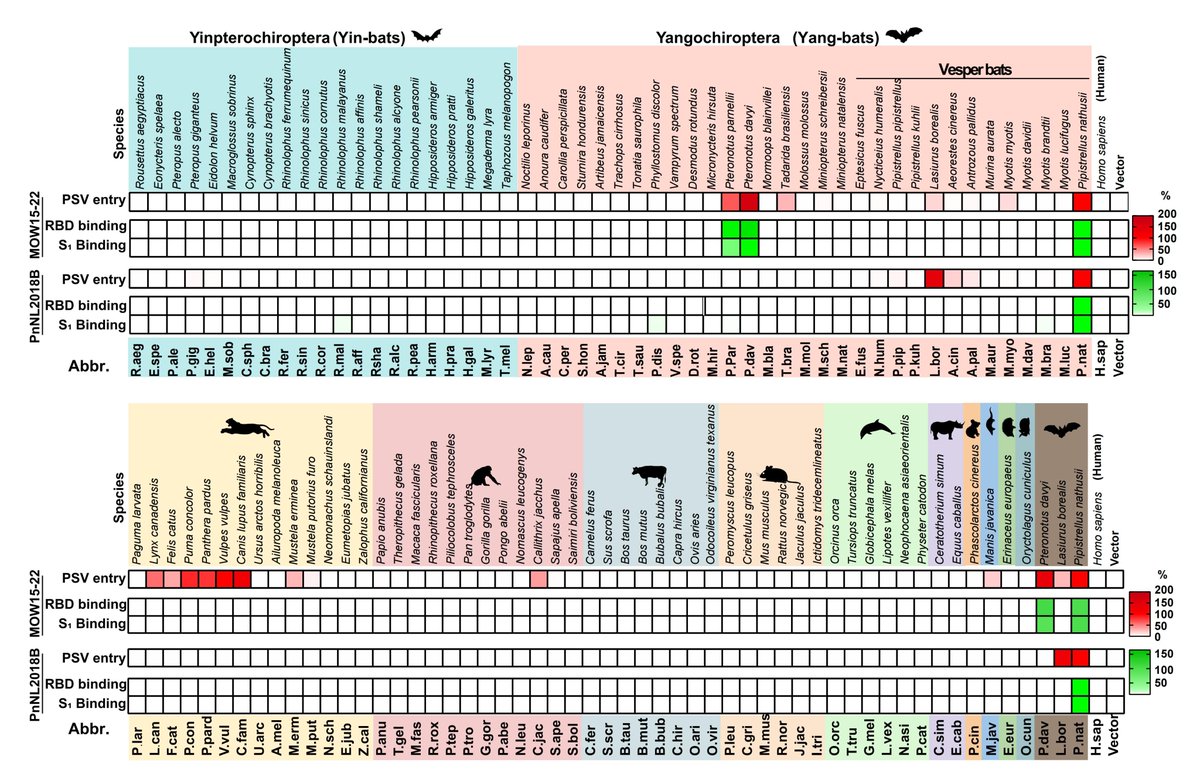
https://x.com/veeslerlab/status/1600540745341227010

https://twitter.com/veeslerlab/status/1730416604029362230






https://twitter.com/yunlong_cao/status/1653804041821093889




https://twitter.com/veeslerlab/status/1473323932388184067

https://twitter.com/veeslerlab/status/1504274269156511745

https://twitter.com/veeslerlab/status/1453109359395762177





https://twitter.com/veeslerlab/status/1476953852767203335

https://twitter.com/veeslerlab/status/1448775318798409729

