#Autism is often associated with an excess of synapses:
but how does this trait affect large-scale circuit function?
Here's what we found by modelling autism-related pruning deficits across-species🧠🐭
➡️tinyurl.com/y2yfj74o
By @MarcoPagani1985 @mvlombardo & al.
Thread 1/n
but how does this trait affect large-scale circuit function?
Here's what we found by modelling autism-related pruning deficits across-species🧠🐭
➡️tinyurl.com/y2yfj74o
By @MarcoPagani1985 @mvlombardo & al.
Thread 1/n

Postmortem investigations in idiopathic #Autism have consistently revealed an excess of excitatory synapses
tinyurl.com/y57cc4r5
tinyurl.com/y59mdffp
tinyurl.com/y3t64yfe
2/n
tinyurl.com/y57cc4r5
tinyurl.com/y59mdffp
tinyurl.com/y3t64yfe
2/n

Seminal work form the #Sulzer_Lab @Columbia has shown that postmortem synaptic surplus in #Autism is associated with hyperactive mTOR signalling
➡️this is a molecular pathway often dysregulated in autism and a key point of convergence of many autism-risk genes
3/n
➡️this is a molecular pathway often dysregulated in autism and a key point of convergence of many autism-risk genes
3/n

This got us wondering: how does this prevalent form of autism synaptopathy affect macro-scale circuit function?
4/n
4/n

As it turns out, mTOR-related synaptopathy can be effectively modelled in the 🐁by knocking out a gene that interacts with mTOR
➡️The gene's name is "Tsc2" and this has been elegantly validated in many previous studies including #Sulzer_Lab
tinyurl.com/yyqaqvqg
5/n
➡️The gene's name is "Tsc2" and this has been elegantly validated in many previous studies including #Sulzer_Lab
tinyurl.com/yyqaqvqg
5/n

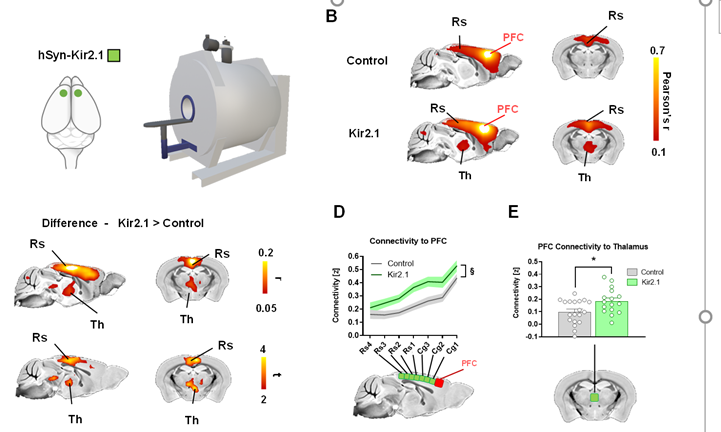
Using fMRI mapping in juvenile Tsc2+/- 🐁 we found that synaptic surplus is associated with fronto-striato-insular hyper-connectivity in these mutants
So this prevalent form of autism synaptopathy DOES affect macroscale circuit function!
6/n
So this prevalent form of autism synaptopathy DOES affect macroscale circuit function!
6/n

We next asked: what could be causing the observed hyperconnectivity?
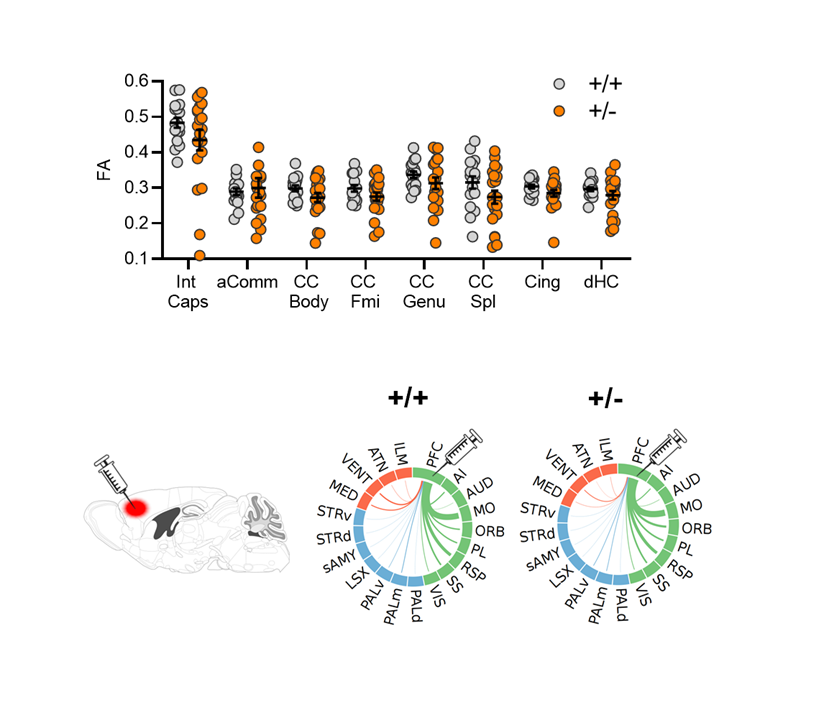
We probed structural connectivity in Tsc2 🐁 but we could not find any gross rewiring or white matter alterations either at macroscale (with DTI) or meso-scale (retrograde viral tracing)
7/n
We probed structural connectivity in Tsc2 🐁 but we could not find any gross rewiring or white matter alterations either at macroscale (with DTI) or meso-scale (retrograde viral tracing)
7/n
Could synaptic surplus possibly *cause* rsfMRI overconnectivity?
In such case, superabundant spines in Tsc2🐭 should be functioning: @RaffaellaTonini probed AMPA/NMDA ratio - a metric sensitive to synaptic maturation - and found that most Tsc2 spines are indeed non-silent
8/n
In such case, superabundant spines in Tsc2🐭 should be functioning: @RaffaellaTonini probed AMPA/NMDA ratio - a metric sensitive to synaptic maturation - and found that most Tsc2 spines are indeed non-silent
8/n

Because spines serve as linear integrators of neuronal input in distributed circuits tinyurl.com/y2mkjo4d,
our hypothesis here is that an excess of spines could lead to increase feedforward connectivity and long-range coupling
9/n
our hypothesis here is that an excess of spines could lead to increase feedforward connectivity and long-range coupling
9/n

To probe the plausibility of this hypothesis @lauraUlysse from the #DecoLab implemented a whole-brain computational model of 🐭rsfMRI connectivity
As predicted, the profile of hyperconnectivity in Tsc2 mice could be modelled by increasing long-range coupling factor "G"
10/n
As predicted, the profile of hyperconnectivity in Tsc2 mice could be modelled by increasing long-range coupling factor "G"
10/n

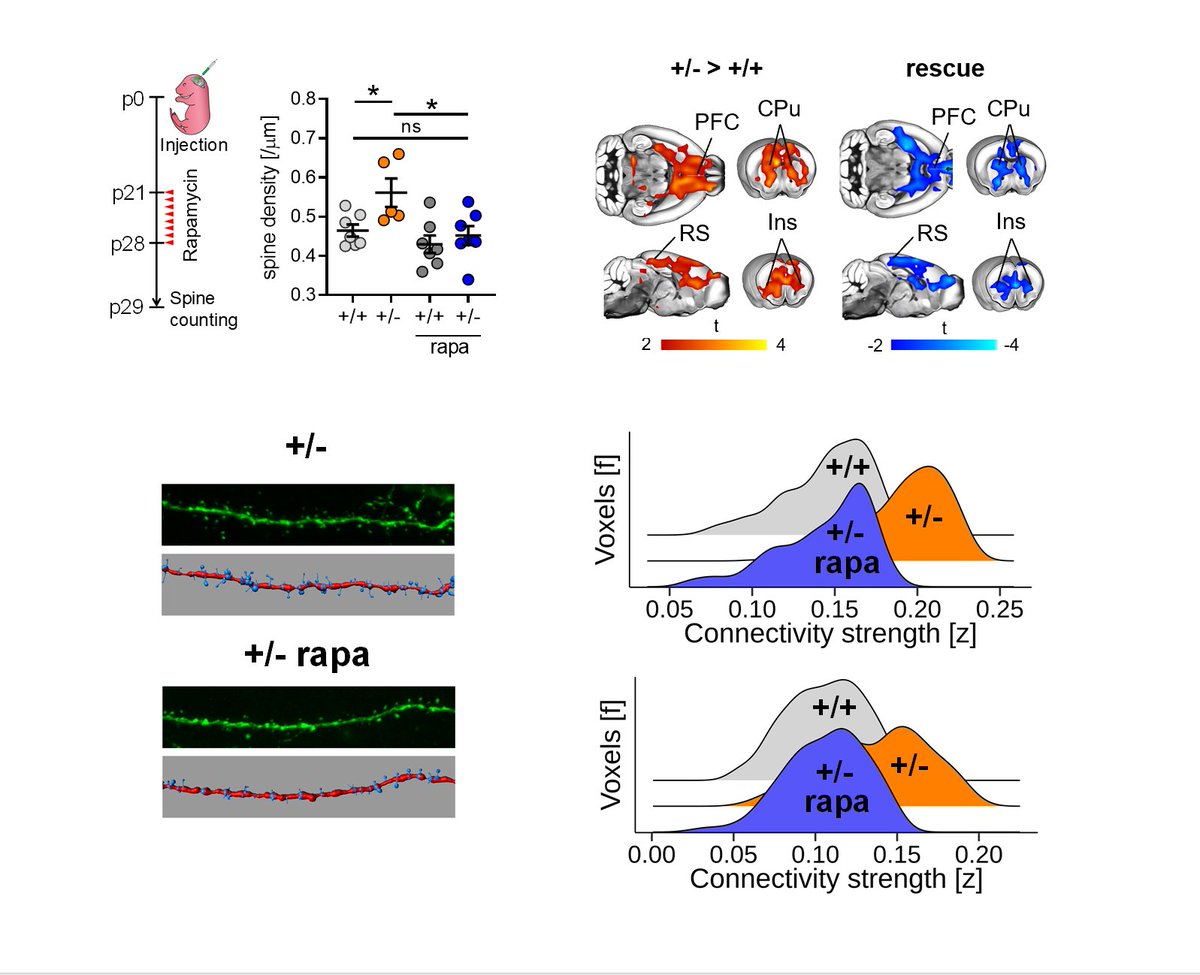
To causally probe a mechanistic link between synaptic surplus and hyperconnectivity we next attempted to normalize mTOR signaling, using rapamycin
This lead to
➡️ complete rescue of synaptic surplus &
➡️ complete rescue of hyperconnectivity
linking the two phenomena!
11/n
This lead to
➡️ complete rescue of synaptic surplus &
➡️ complete rescue of hyperconnectivity
linking the two phenomena!
11/n
Interestingly, we also found that
➡️Social deficits and increased stereotypies in Tsc2 🐭 are completely rescued by rapamycin
➡️ fronto-cortico-striatal hyper-connectivity is a good predictor of motor stereotypies!
12/n
➡️Social deficits and increased stereotypies in Tsc2 🐭 are completely rescued by rapamycin
➡️ fronto-cortico-striatal hyper-connectivity is a good predictor of motor stereotypies!
12/n

The results of these 🐭 investigations are exciting because they mechanistically reconcile autism synaptopathy & connectopathy within a unifying multi-scale framework.
But are they clinically relevant?
13/n
But are they clinically relevant?
13/n
The high prevalence of synaptic surplus and mTOR hyperactivity in idiopathic autism suggest that a similar hyperconnectivity signature could be identifiable in patients.
We thus examined fMRI scans from ABIDE-I and found clear hotspots of hyperconnectivity in insular areas
14/n
We thus examined fMRI scans from ABIDE-I and found clear hotspots of hyperconnectivity in insular areas
14/n

We next probed the corresponding networks involved and found the children with autism exhibit fronto-striato-insular overconnectivity, like observed in Tsc2 🐭!
This is consistent with our hypothesis, but how can we link this signature to mTOR signalling?
15/n
This is consistent with our hypothesis, but how can we link this signature to mTOR signalling?
15/n

To do that, we run a gene decoding analysis of the observed hyperconnectivity signature
This analysis revealed that the identified fronto-striato-insular signature is significantly enriched with genes interacting with mTOR and TSC2!
16/n
This analysis revealed that the identified fronto-striato-insular signature is significantly enriched with genes interacting with mTOR and TSC2!
16/n

However autism is heterogenous and not exclusively mTOR-related!
So the observed group-level association MUST be driven by a specific subgroup of patients in which this signature is especially prominent
To test this hypothesis we clustered insular connectivity profiles..
17/n
So the observed group-level association MUST be driven by a specific subgroup of patients in which this signature is especially prominent
To test this hypothesis we clustered insular connectivity profiles..
17/n

...and found:
➡️four distinct connectivity profiles (autism is INDEED heterogeneous!)
➡️as predicted only one subtype (#2) exhibits highly enriched mTOR interactome expression --> it drives group level changes!
18/n
➡️four distinct connectivity profiles (autism is INDEED heterogeneous!)
➡️as predicted only one subtype (#2) exhibits highly enriched mTOR interactome expression --> it drives group level changes!
18/n

So we causally linked mTOR-related synaptic pathology to large-scale circuit alterations, and identified a putatively segregable novel autism subtype!
Very grateful to @SFARIorg for generous funding & @StavrosTrak @LabPasqualetti @alibert_ & all twitterless collaborators!
Very grateful to @SFARIorg for generous funding & @StavrosTrak @LabPasqualetti @alibert_ & all twitterless collaborators!
• • •
Missing some Tweet in this thread? You can try to
force a refresh