Our paper on the role of environmental factors on #SARSCoV2 transmission (led by Canelle Poirier & @MauSantillana) has been peer-reviewed & is now live: nature.com/articles/s4159…
In our study, we find a lack of sufficient evidence for weather-mediated seasonality of #COVID19. 1/X
In our study, we find a lack of sufficient evidence for weather-mediated seasonality of #COVID19. 1/X
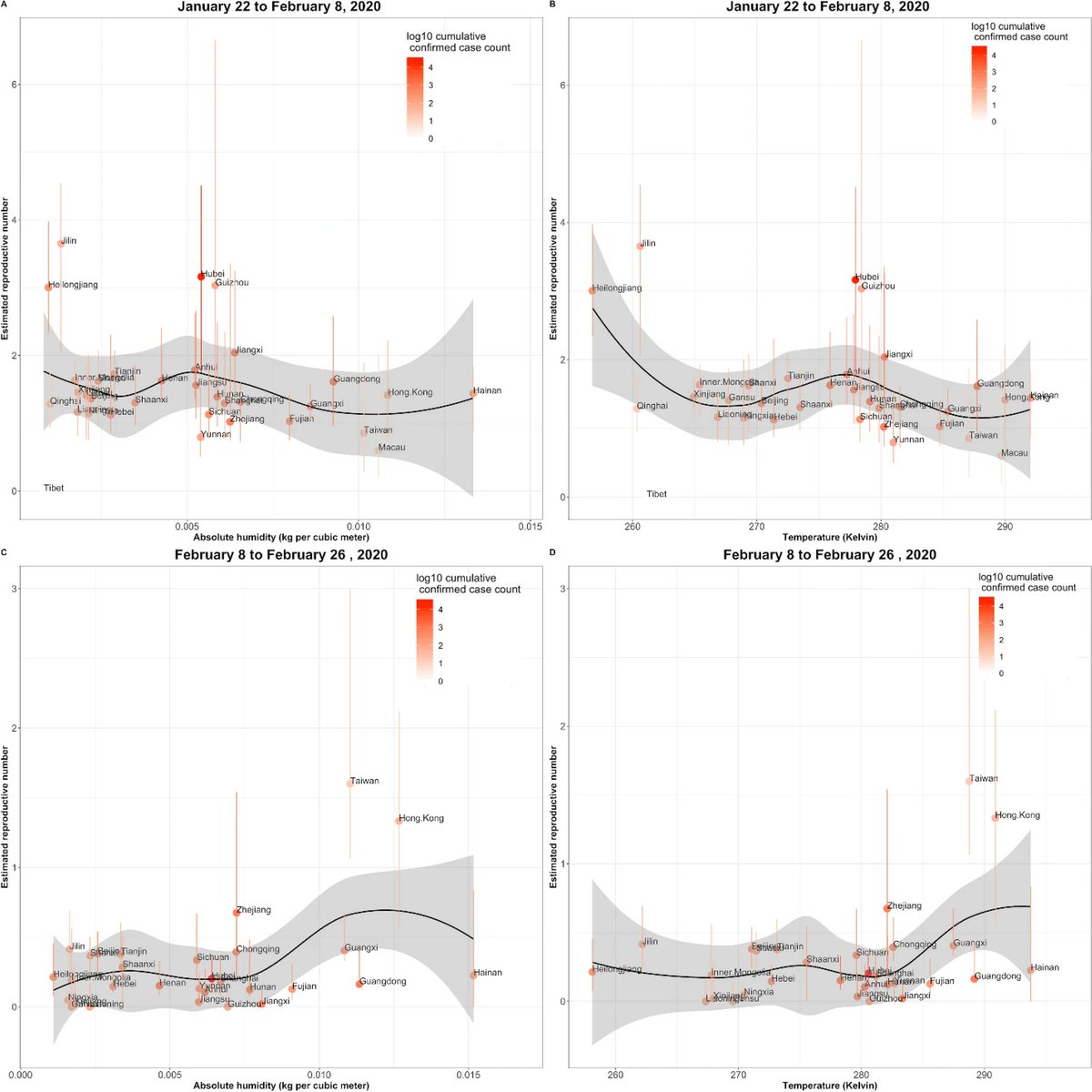
I’ll let @MauSantillana take the lead on fielding questions about this work, but I wanted to point out two things.
First, I use the term “weather-mediated seasonality” conscientiously. There are a lot of other phenomena that *also* mediate seasonality, like behavior change. 2/X
First, I use the term “weather-mediated seasonality” conscientiously. There are a lot of other phenomena that *also* mediate seasonality, like behavior change. 2/X
For example, I don’t doubt that congregating indoors – which will likely happen more often as winter emerges in the U.S. – may very well increase transmission risk in temperate climates.
These issues are extremely important, but they aren't what we’re studying in this paper. 3/X
These issues are extremely important, but they aren't what we’re studying in this paper. 3/X
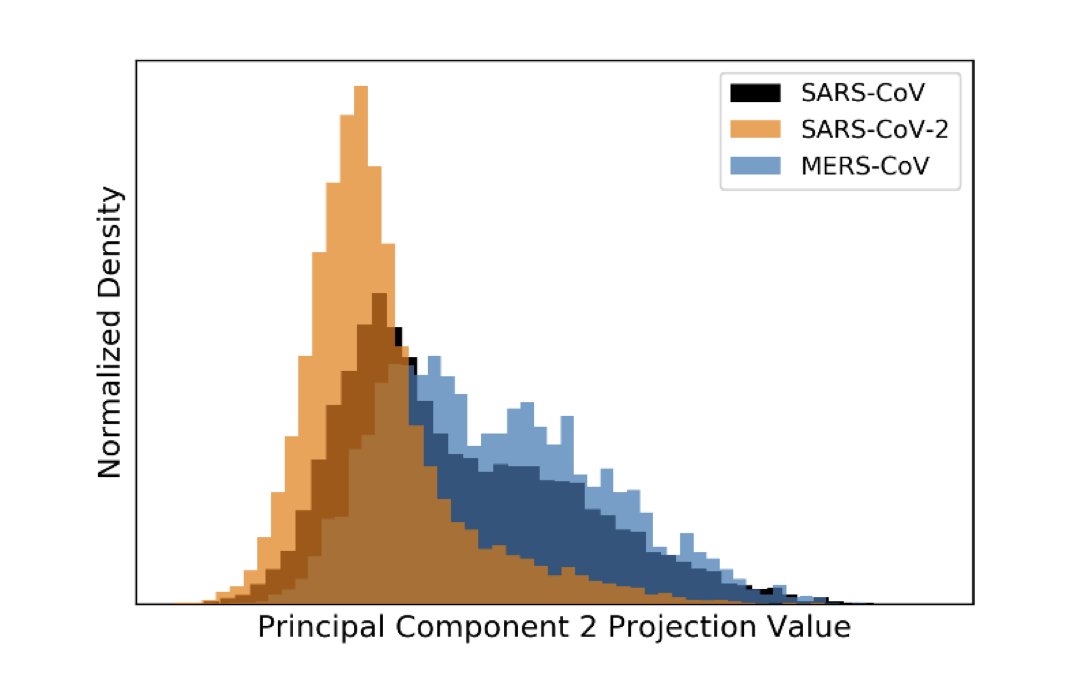
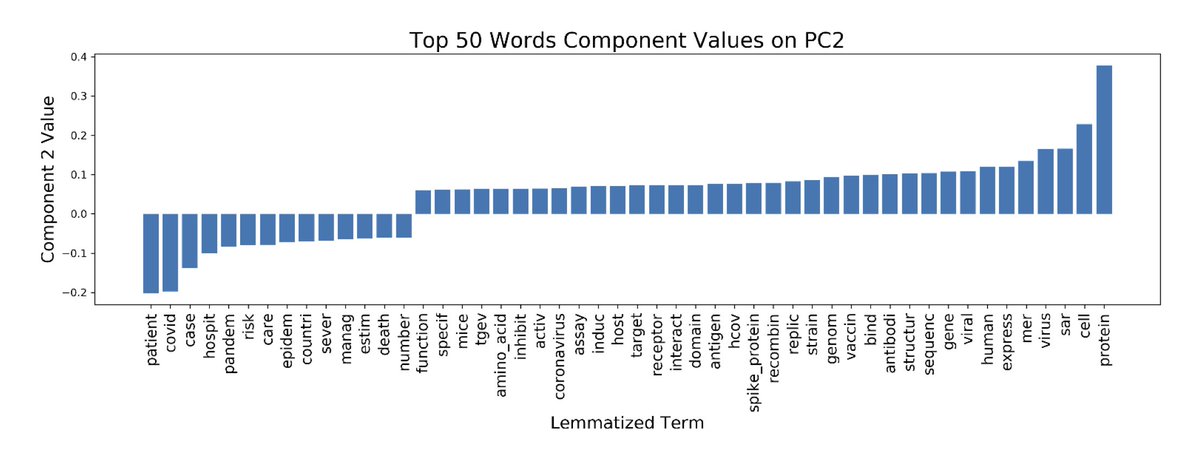
Instead, we examine the spatial variability of #SARSCoV2 transmission across China & show that weather-related (or environmental) factors alone can't explain said variability. From there, we draw conclusions that are now unsurprising, given summertime upticks in #COVID19. 4/X
Which brings me to the second point I want to make today.
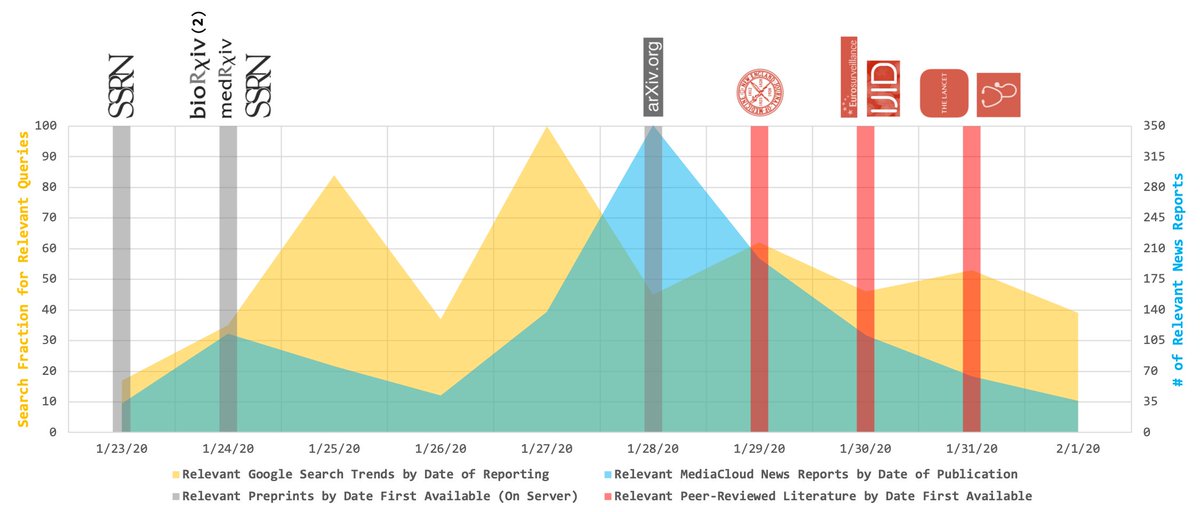
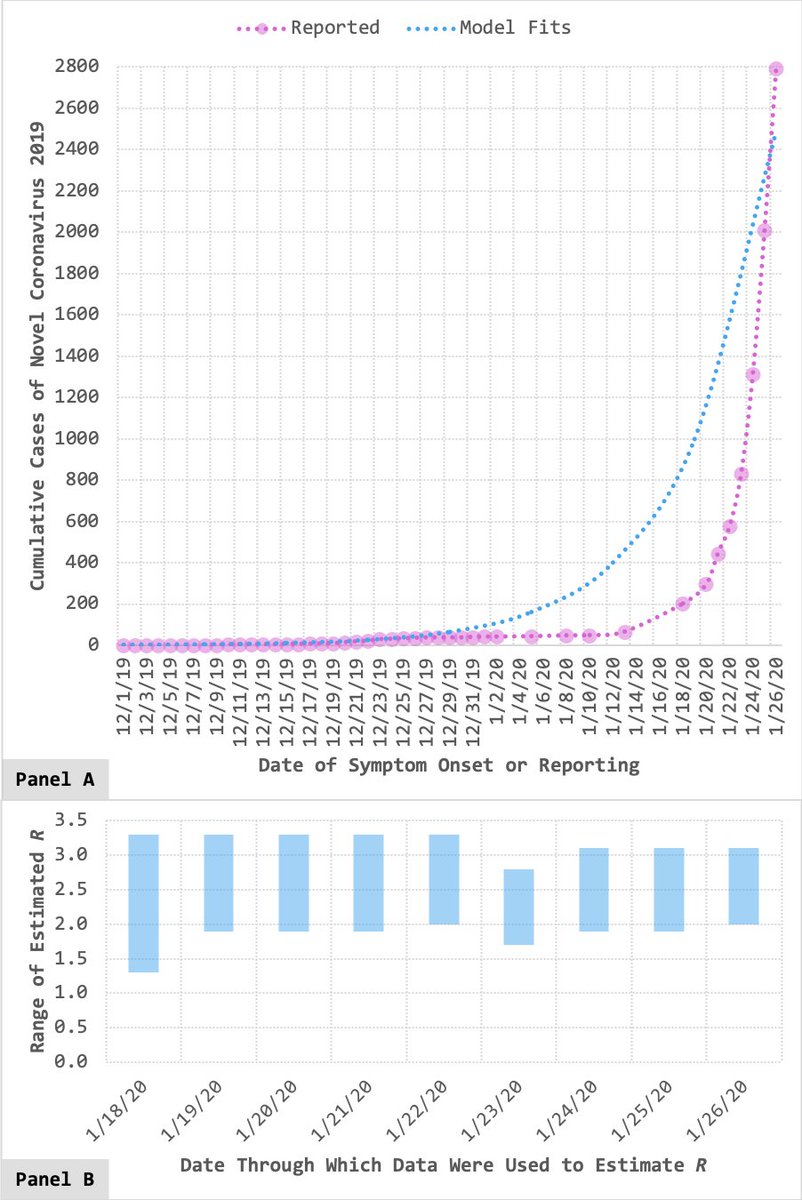
This paper was first posted as a preprint in March 2020 (papers.ssrn.com/sol3/papers.cf…). At the time, summer hadn’t yet arrived in the Northern Hemisphere (which made us the bearers of bad news, considering our findings). 5/X
This paper was first posted as a preprint in March 2020 (papers.ssrn.com/sol3/papers.cf…). At the time, summer hadn’t yet arrived in the Northern Hemisphere (which made us the bearers of bad news, considering our findings). 5/X
…But if we hadn’t posted our work as a preprint, this study wouldn’t have been released to the public until today – well-past summertime in places like the U.S. Thanks to preprinting, we were able to share what we’d found early enough to (potentially) make a difference. 6/X
Whether or not we actually *made* a difference is a separate story... But to me, this is the beauty of preprinting. Though preprinting has serious weaknesses (e.g., it can prompt the proliferation of studies that are largely unvetted), it also allows us keep science current. 7/X
And with that, I'll wrap up with a thank you to @MauSantillana for letting me be part of this important study. That said, our work (like all science) has limitations (many of which are noted in the manuscript) – so if there are any curiosities, please direct them his way! 8/8
• • •
Missing some Tweet in this thread? You can try to
force a refresh