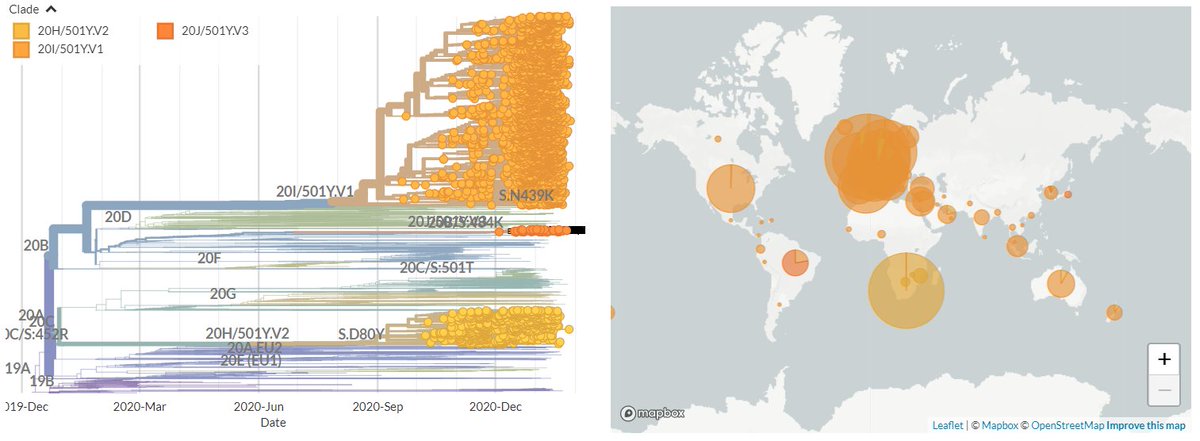
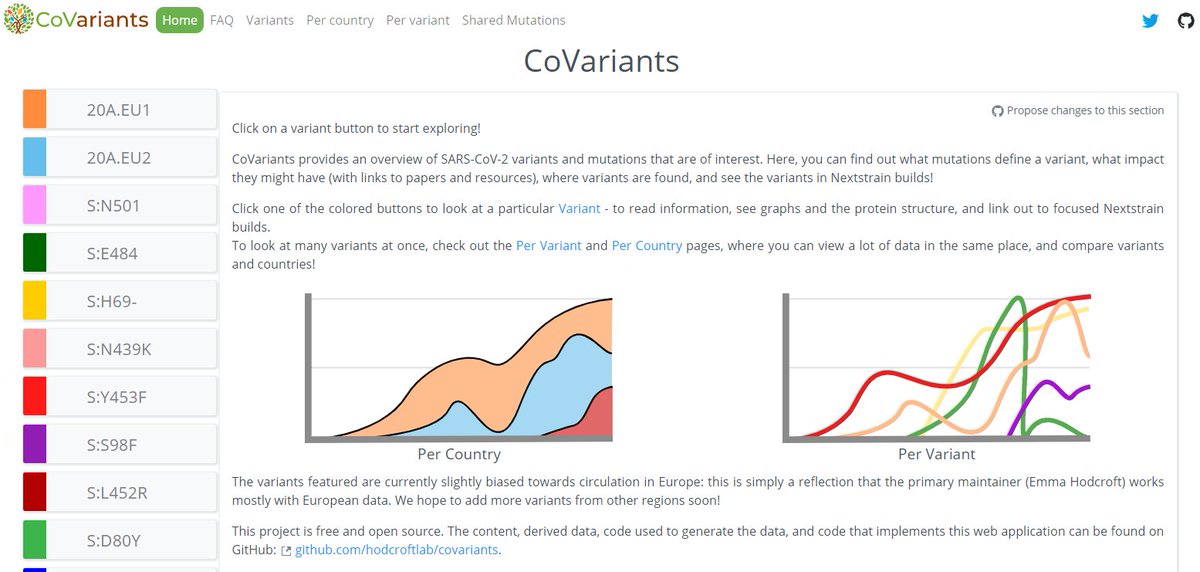
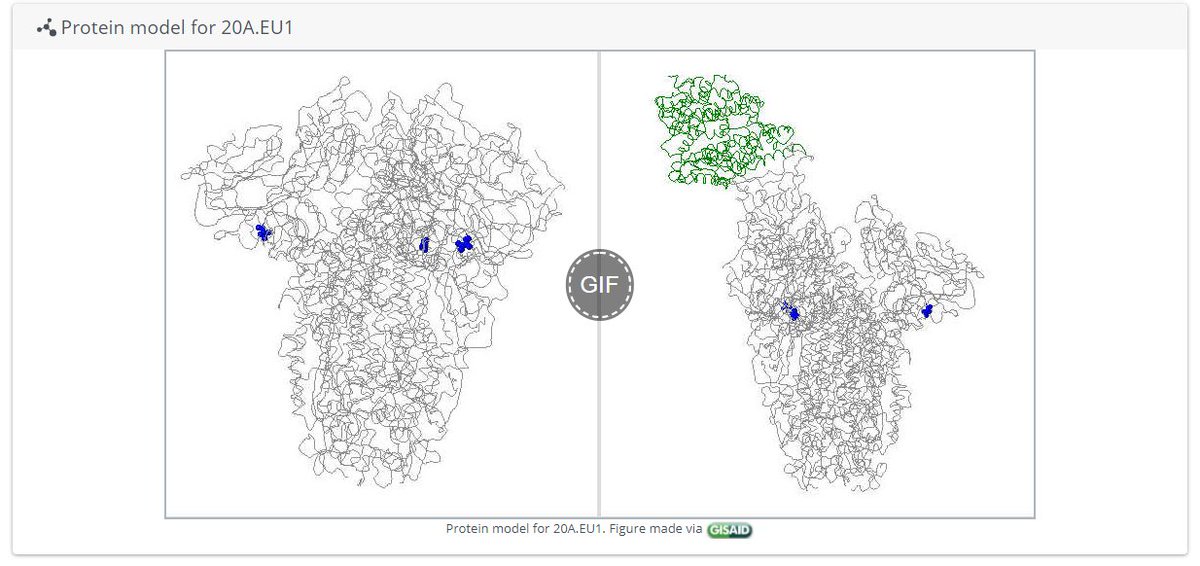
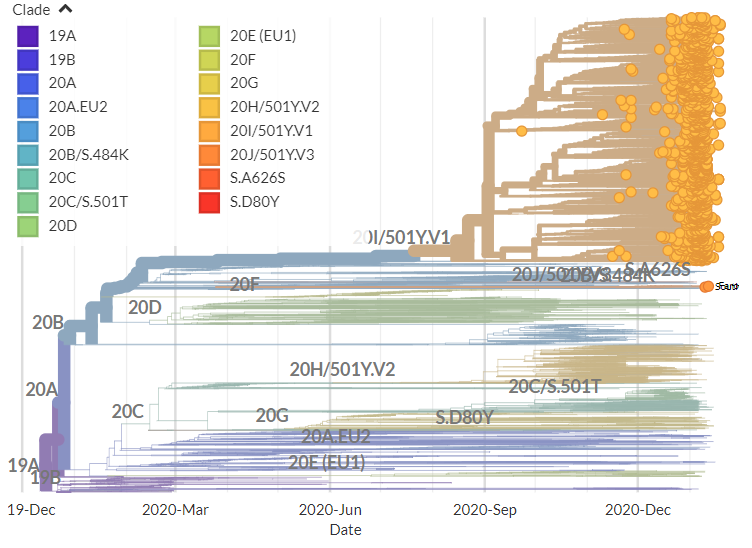
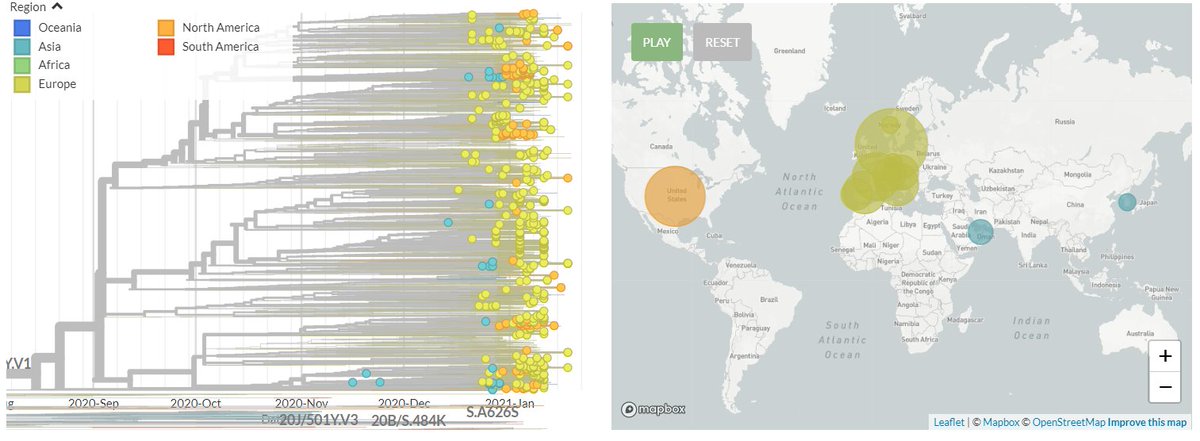
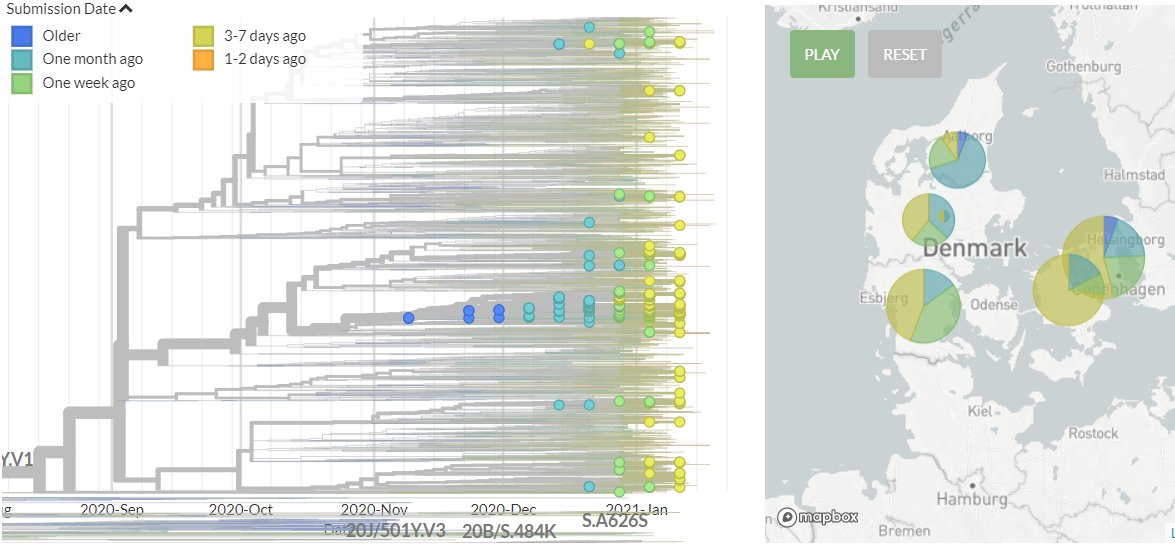
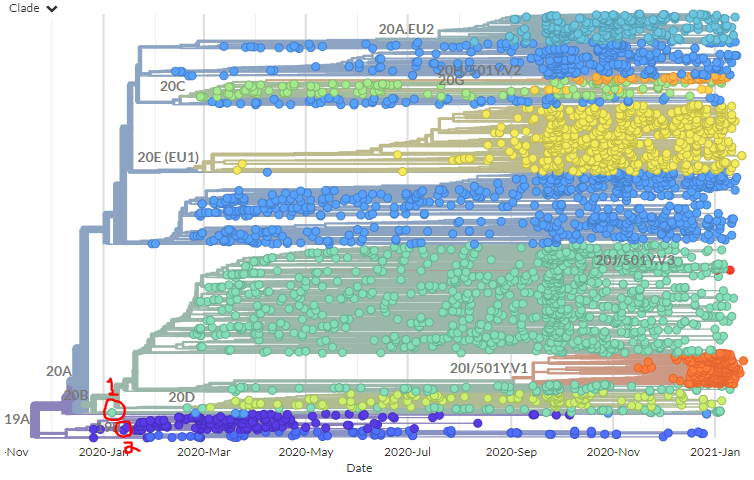
I'm still getting a *lot* of media enquiries - more than I can even respond to! So here's some of my best tips for how to reach out to me to maximise your chances of success (though I imagine they're true of other scientists too!)
1/7
1/7
(Just as a disclaimer - even if you use all of these, I can't promise I write back, or that I can say yes. If emails slip off the 'front page' of my inbox they basically fall into a black hole 😬. And sometimes I just cannot take any more interviews, even great ones.)
2/7
2/7
1. Please tell me what time zone you're in!
If I want to say 'yes' that means a pencilled-in appointment goes into the calendar immediately - so I need to know when you're likely to be awake. If I have to email you to find out... it might not happen.
3/7
If I want to say 'yes' that means a pencilled-in appointment goes into the calendar immediately - so I need to know when you're likely to be awake. If I have to email you to find out... it might not happen.
3/7
2. Please tell me your deadline!
I don't mean your editors deadline or publishing deadline. I mean - do you need to talk to me in 24 hours? By Friday? By next Wednesday? Again, this means I can pencil this in my calendar *straight away* (or at least let you know it's a no).
4/7
I don't mean your editors deadline or publishing deadline. I mean - do you need to talk to me in 24 hours? By Friday? By next Wednesday? Again, this means I can pencil this in my calendar *straight away* (or at least let you know it's a no).
4/7
(Tip: the more time you give me the better my chance of response. I know that isn't always in journalists' control, but my days fill up fast and last-minute things are often impossible to squeeze in.)
5/7
5/7
3. Stand out from the crowd!
I get a lot of the same emails every day: "I'd like to talk to you about the variants."
Why should I pick email 1 over email 2? Ask an interesting question. Show you've done your research. Make me like your email better!
6/7
I get a lot of the same emails every day: "I'd like to talk to you about the variants."
Why should I pick email 1 over email 2? Ask an interesting question. Show you've done your research. Make me like your email better!
6/7
4. Make it impossible for me not to respond!
Ask me the question nobody's asked me yet & your chances of getting a response shoot up. I spend a lot of my media time repeating myself. Give me a chance to take a new angle or a subject I care a lot about - I can't stop myself!
7/7
Ask me the question nobody's asked me yet & your chances of getting a response shoot up. I spend a lot of my media time repeating myself. Give me a chance to take a new angle or a subject I care a lot about - I can't stop myself!
7/7
• • •
Missing some Tweet in this thread? You can try to
force a refresh