
My highlights of the RNGS21 @MGI_BGI Satellite Workshop talks: vibconferences.be/events/revolut…
Talk on AMR genes by @johnpenders at Maastricht UMC+ (I hope it's the right one), and how these increase/decrease for example with intercontinental travel patterns. Travelers to South-Eastern Asia acquire the mcr-1 gene. 
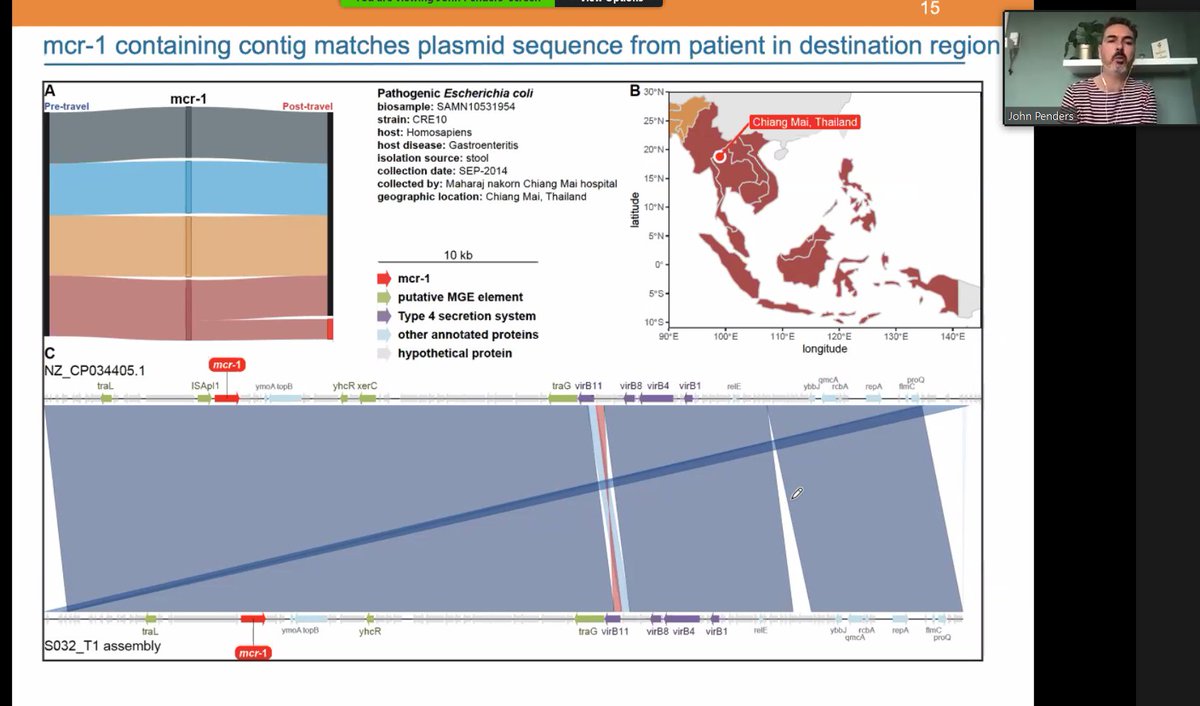
Mcr-1 gene is identified and well-known from 2015, but the patterns of AMR migration started earlier 

My conclusions: very similar parallel patterns between the acquisition of AMR genes with travel and the spread of #COVID19 variants across the globe: the former doesn't kill millions of people but the #NGS technology to track them is very equivalent. 



Next talk Prof. Miguel Esteban, CAS: single-cell transcriptomic atlas of Macaca fascicularis. Showing the large factory-like DNBSEQT10 in the picture 

On the #singlecell side, they are already running the @MGI_BGI DNBelabC4 instruments (@SanDiegOmics they do exist) 



Co-expression of ACE2 with IL6R/IL1R1/IL1RAP (cytokine response/storm) for #COVID19 #singlecells #DNBelabC4 
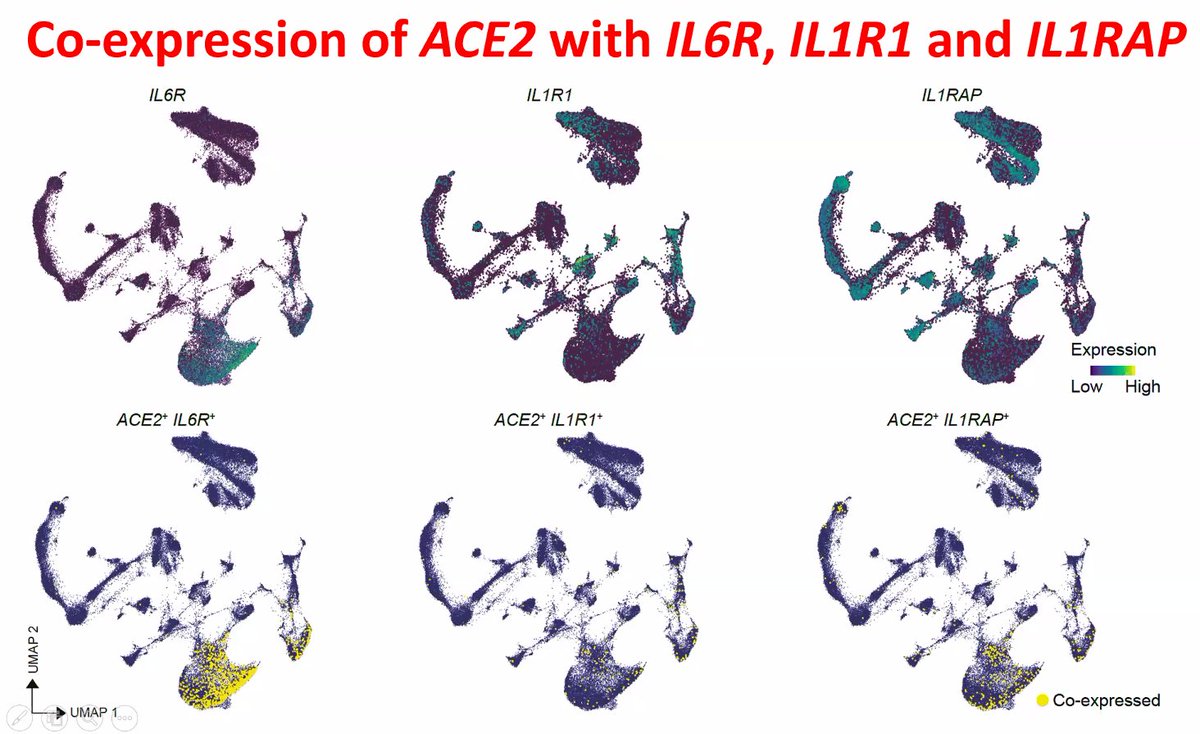
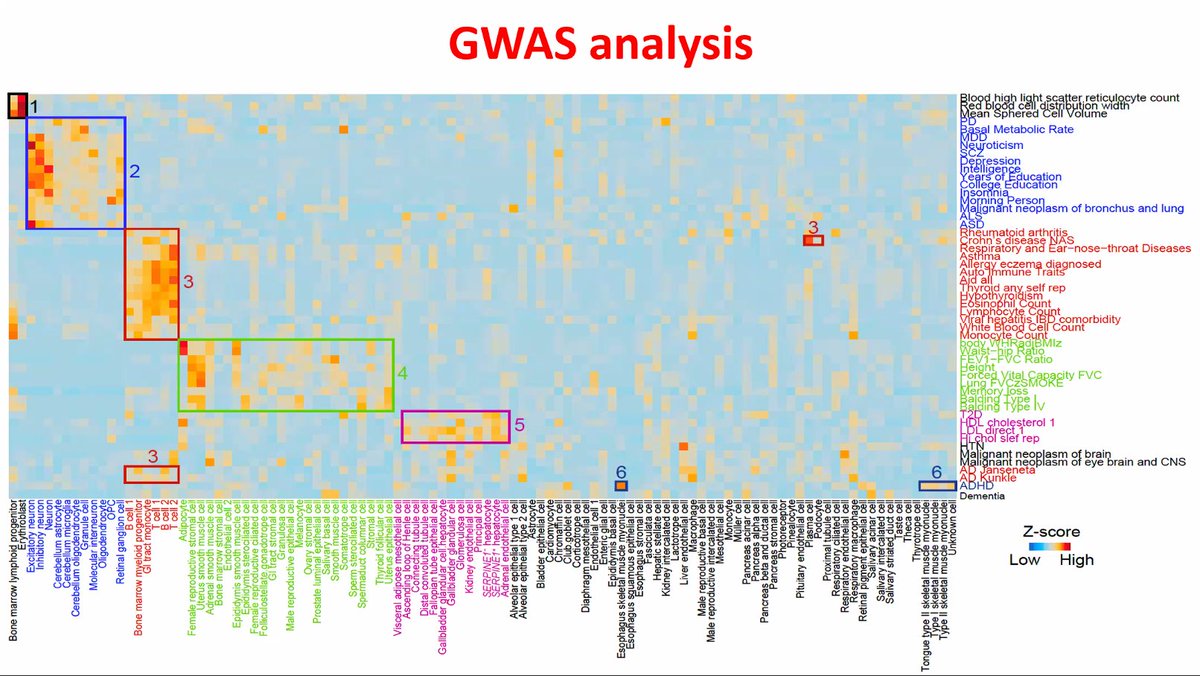
Older people overreact to #SARSCoV2 infection in a cytokine storm that affects prognosis. Tocilizumab brings down storms to manageable levels. Correlates with genome-wide markers in blood cells but also surprisingly ADHD. 


Q. How does DNBelabC4 compare to @10xGenomics Chromium?
A. "Ours is at least as good"
#singlecell bit.ly/scellmarket
A. "Ours is at least as good"
#singlecell bit.ly/scellmarket
Next talk is from Dr. Ao Chen, BGI Group: "Large field of view-spatially resolved transcriptomics at nanoscale resolution"
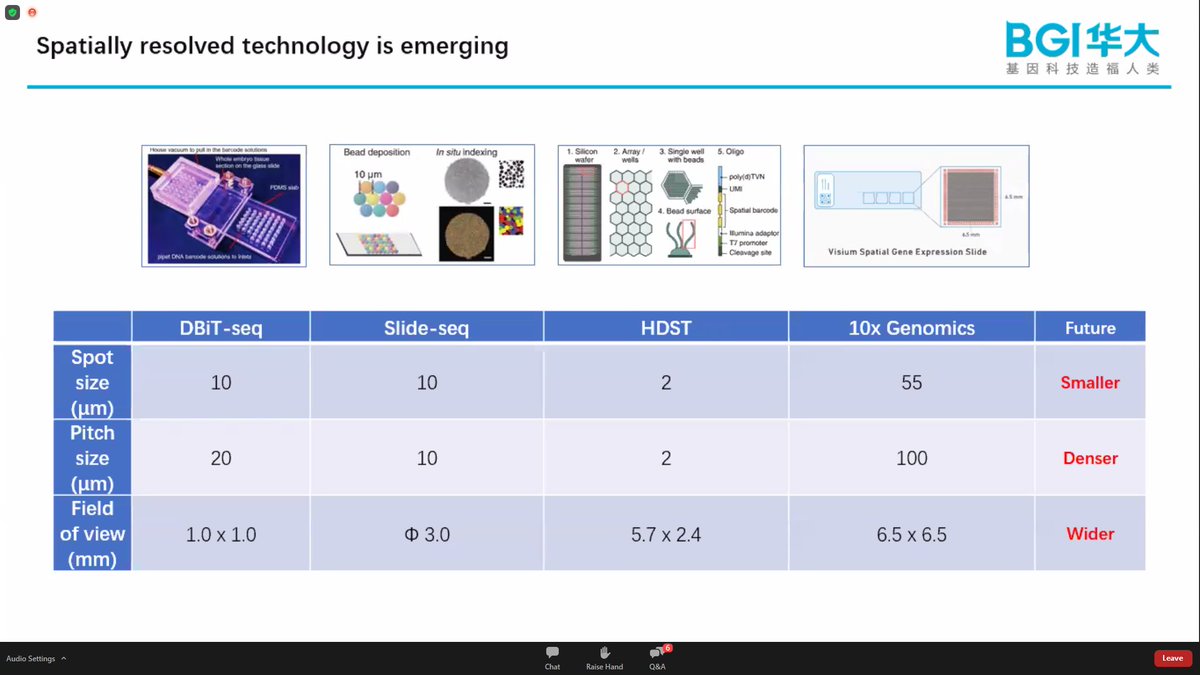
Spatio-Temporal Enhanced Resolution Omics-Sequencing (Stereo-seq) at nanoscale resolution - Ao Chen. Current methods include bit.ly/scspatial 
The 6-step stereo-seq method at @MGI_BGI uses the same nano ball technology as the #DNBSEQ technology *and* the same type of flowcells. The slide can be as large as the flowcell (i.e. massive). 

Specs of @MGI_BGI stereo-seq : the size of the palm or a monkey brain. Subcellular localization (500nm / 0.5um) 

Comparing DNB blood vessels of different sizes vs 100um resolution (right). Example mouse embryo (16.5 days). 



Introns can be subcellular localized within the cell nucleus. Some of the introns can be secreted outside of the nuclei, but the majority stay (as textbooks say) inside the nuclei. 

Very large FOV and resolution means billions of nanoballs which can be analysed in 3D within the slice. Digital pathology (liver cancer) applications. 



More applications to come, some demoed on this website: stereomap.cngb.org 

@d2unroll unroll
• • •
Missing some Tweet in this thread? You can try to
force a refresh










