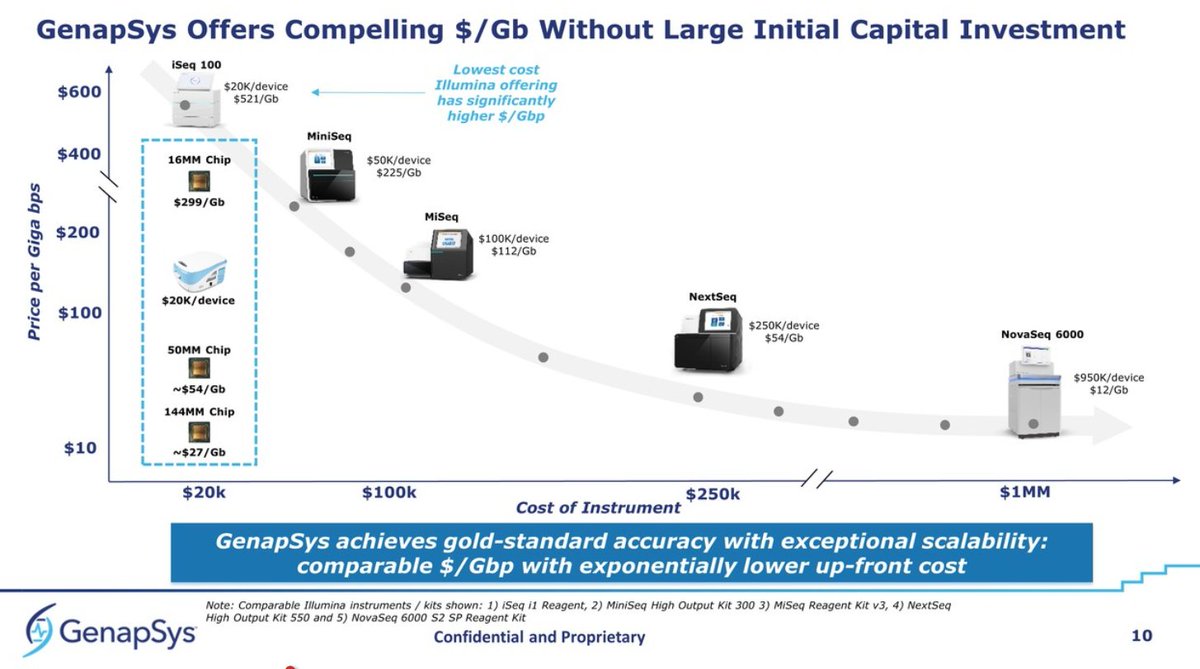
Epigenomic biomarkers are becoming more established for #LiquidBiopsy and #CancerScreening, and we have seen the big players positioning themselves in this #epigenomics race recently.
Catching signs of #cancer early is crucially important to the disease management and survival rates, so the question is: how can we find out if there is something wrong going on early enough, ideally in a low-cost assay that can be performed regularly on healthy individuals?
The first generation of high-throughput technologies (NGS) was predicated on finding mutated (tumor) DNA in the individual's body (somatic), different from their (normal) DNA. Tumor/Normal (T/N) comparisons of the individual's samples, biopsies or ctDNA will indicate if there
are signs of cancer already underway. So why do we need #epigenomic profiling if we can do all this with the detection of mutations? Well, what scientists found is that if you can see sufficient mutated copies of DNA, cancer may already have progressed beyond the initial stages.
Then scientists asked the question: is there an earlier mark that we can detect for earlier stages of cancer before the Tumor/Normal mutations signal raises up? Here is where #epigenomics profiling started playing a role.
Out of the big players, $GH Guardant Health was one of the early adopters of #epigenomic profiling for early cancer detection. The slide decks of their R&D projects mentioned #methylation (5mC) as a way of catching cancer earlier than with mutation profiling.
Later on, $GH brought in #fragmentomics via a combination of their own developments and acquisitions. The fragmentomics profiling of ctDNA is relatively easy and complementary to #methylation marks and adds up to better AUCs.
While $GH was working on this,
@GrailBio came to the fore with a big couple of funding rounds, the backing of @illumina, and a very clear mission statement to go after cancer screening -- I don't think it has changed since then.
@GrailBio came to the fore with a big couple of funding rounds, the backing of @illumina, and a very clear mission statement to go after cancer screening -- I don't think it has changed since then.
At @GrailBio, they looked at different options, and again #methylation profiling became the tip of the arrow. I haven't followed the IP stemming from their internal work, but in slide decks & bits of PR, there is the constant mention of their bisulfite-free method to assay 5mC.
That's an important consideration for #LiquidBiopsy as there isn't *that* much DNA present in a 10ml draw of blood -- the size of a lipstick. There could be 500-1000 copies of the genome, most in fragments of around 150-160bp. So old school methods like bisulfite assays simply
destroy most of the fragments composing these 1000 copies in a #LiquidBiopsy sample, so you end up with not enough left to have a good signal-to-noise ratio. The final profile is ideally something like this screenshot (from TwistBio material): 
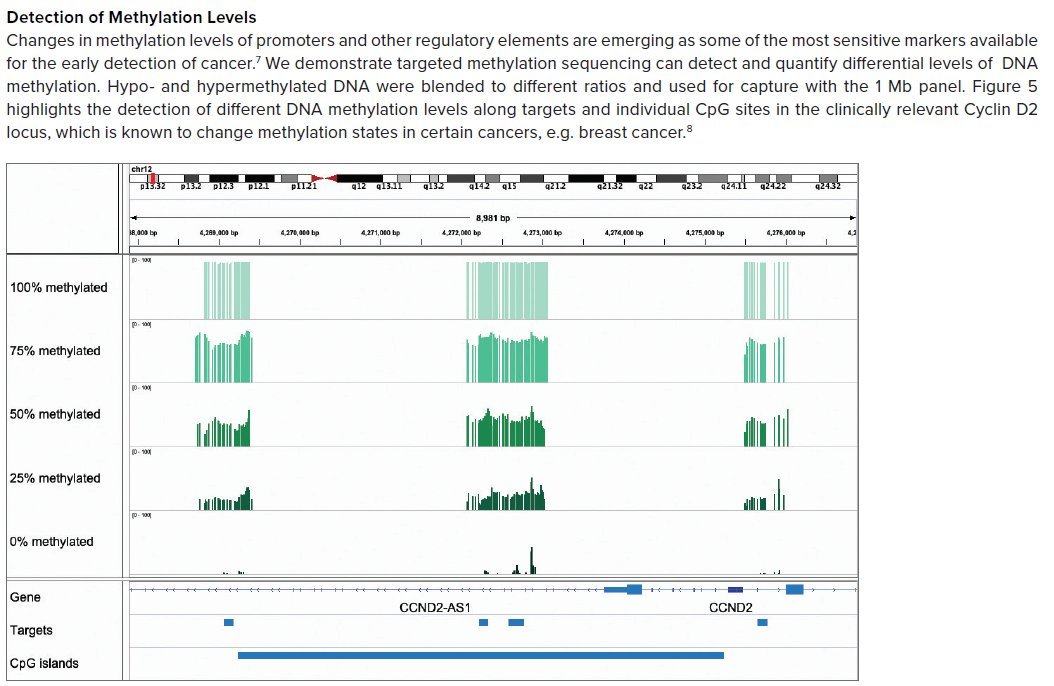
where the peaks show places of presence of 5mC: the highest the peak, the higher the fraction of all copies is methylated. The places without peaks are almost always unmethylated (5C). This binary 5mC vs 5C switch for each CpG in the genome is the basis of the biomarker.
So what's the ideal? A method that takes the smallest amount of blood, takes into account the largest amount of genome copies (coverage) without destroying them during the assaying, and also ideally, that can find haplotypic information: long stretches of the genome where
the signal is "linked" (in linkage) for several of these CpGs. In terms of coverage, the more you sequence the higher depth that will be assayed, and the bigger the panel of loci, the higher the throughput needed to cover it more deeply. This has a cost consideration, as the more
you sequence, the more you have to pay either
@illumina or @MGI_BGI or other short-read technology for a given profile. Long reads are of no benefit "per see" in #LiquidBiopsy #cfDNA, as the fragments are short in the bloodstream. If your method destroys part of the DNA while
@illumina or @MGI_BGI or other short-read technology for a given profile. Long reads are of no benefit "per see" in #LiquidBiopsy #cfDNA, as the fragments are short in the bloodstream. If your method destroys part of the DNA while
assaying it, then you'll end up with a less diverse pool to start with, and more of your NGS throughput will be wasted in duplicate reads (reading the same sample fragment more than once).
So what's out there that's better than bisulfite 5mC? Both $GH and @GrailBio claim to have their own method. Then $EXAS acquired for a stonking $515M a company, #BaseGenomics that has a method to do this, both for 5mC or 5hmC (a related but different epigenetic mark to 5mC).
Then there is the De Carvalho lab who does this differently: if you need to have this signal of 5mC peaks, and most of them are going to be consistently the same for about 50-60% of the genome, why not instead of sequencing 100% of the genome, do an enrichment method that
gives you as many reads as the height of the peaks when they are methylated? This way, in the binary 1/0 signal, for 5mC/5C, you only spend your throughput sequencing the 1s, and don't spend it with the 0s. Then you extrapolate the 0s with the difference of the peak heights.
This idea is great in principle but has been difficult to implement for small amounts of DNA. The De Carvalho lab, and their spin-off DNAmx (tdc.uhnresearch.ca/sites/default/…) has shown progress on the original work that was done by the Reik Lab in Cambridge UK.
So if pulling down the 5mC fraction of a cfDNA sample is cheaper because it concentrates the throughput in the 50-60% of the genome that is consistently methylated, then what else can be done to further drive down the cost of each #epigenomic profile?
One way is to do this in a targetted fashion: only select loci that you think are interesting as part of the biomarker. @GrailBio solved this with the "brute force" approach: spend lots of money doing it genome-wide, then distill the targetted panel from it.
In this way, we take out the "boring" part of a 5mC profile and only include the regions that could maximally inform a healthy vs cancer diagnosis. Another approach is to find a different epigenomic mark that has less of the "boring" part, and not have to design a panel.
Here is where 5hmC is now shining new light in early cancer diagnosis. The 5-hydroxymethylation marks are concentrated into 8-12% of the genome, into what people would class as functionally enriched parts of the genome, such as genes (gene bodies), promoters, proximal enhancers
and other functional elements of the genome. This difference between 5hmC and 5mC, one being concentrated at around 8-12% of the genome and the other at 50-60% is due to a historical event in the genome of most plants and animals: the introduction of repetitive DNA in genomes.
Many millions of years ago, the genomes of eukaryotes (not so much bacteria and archaea) started to accumulate repetitive DNA, in many cases of foreign origin: viruses and other vectors would infect you and in some cases, they would leave a trace of DNA inserted in your genome.
The solution for bacteria and archaea, which need to divide very quickly and efficiently, was to have a defense mechanism to chop off this external invasive DNA, and keep their genomes nice and compact. The CRISPR technology itself has an origin in these attempts to protect your
genome from viruses that insert their DNA onto your genome. But for plants and animals, the solution was to not try to get rid of this repetitive DNA, but at least keep it under control. And one way of doing that was to paint it with the methylation brush: repetitive methylated
DNA would then not interfere with the normal functional workings of the genome, and plants and animals thus adapted to cope with repetitive DNA in their genome. And here lies the difference between the 8-12% of the genome that may contain 5hmC and the 50-60% of the genome that
usually contains 5mC. This boring repetitive methylated DNA is now an issue in trying to drive down costs for assays, while at the same time, we want to assay as genome-wide as possible so that we capture all functional elements of the genome.
So in early cancer diagnosis, there are now companies applying the same theory of 5mC biomarkers to the 5hmC profiling.
enseqlopedia.com/2020/06/brain-…
enseqlopedia.com/2020/06/brain-…
How about combining signals? E.g. would, in theory, a 5mC+5hmC profile have a better AUC than either in isolation? How about adding fragmentomics? How about proteomics? How about CTCs? You name it.
In theory, yes, the wider scope of your signal, the better the specificity and sensitivity of the diagnostics assay. In practice, also yes, but with caveats: you could make the perfect early cancer diagnosis with a combination of all current technologies and future ones, but it
would take such a long time to train the model on a humongous training data set that encompassed enough samples to have the breath of particular cases for the training. So cutting back on the less important signals means you can then practically have better
AUC than any single assay and still be able to practically train the model and then apply the classifier on the next sample that comes through the door.
So what will be the future best assay for #LiquidBiopsy #cancer #screening?
So what will be the future best assay for #LiquidBiopsy #cancer #screening?
In my opinion, we are not too far from having a multisignal (multiomics) assay that encompasses a deep 5mC profile, in breadth and coverage), a deep genome-wide 5hmC profile, and a shallow sequencing fragmentomics profile. This will take more than 10ml of blood, but could be 20ml
and the technology is there for that. What else we can add to it? CTCs? Proteomics? I'll leave it to the comments for people to chip in.
@d2unroll unroll
• • •
Missing some Tweet in this thread? You can try to
force a refresh