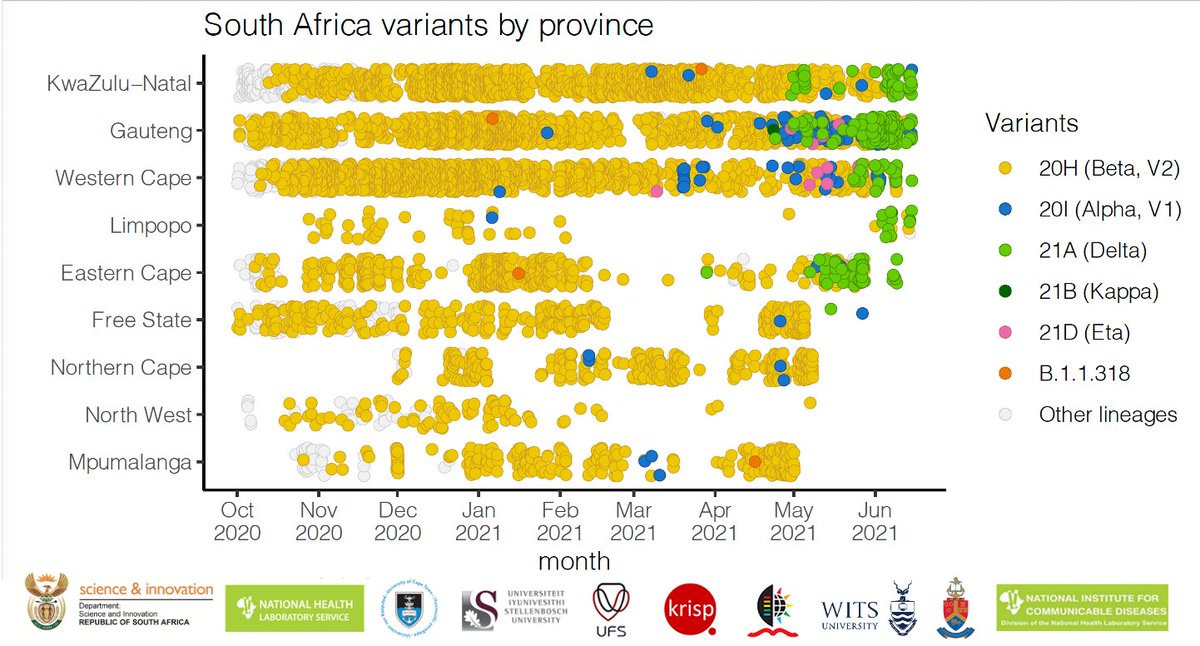
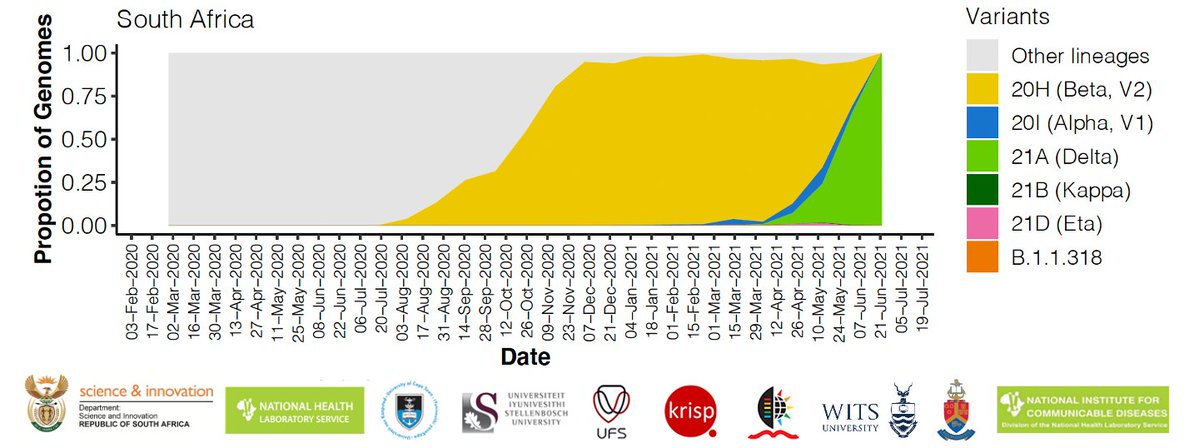
NGS-SA worked hard to increase sequencing in South Africa. We know now that the explosive #COVID19 third wave is dominatedd by the Delta variant.
In this tweet thread, we present recent data (sampled up to 24 June in many provinces in SA, including Gauteng) - Tweet 1/8
In this tweet thread, we present recent data (sampled up to 24 June in many provinces in SA, including Gauteng) - Tweet 1/8
The third wave in Gauteng was caused by the Delta variant, thanks to @nicd_sa and @krisp_news who worked day and night on the weekend to increase genomics surveillance in Gauteng.
Graph shows Black line = Daily cases (7-day average), Yellow = Beta, Green = Delta. Tweet 3/8
Graph shows Black line = Daily cases (7-day average), Yellow = Beta, Green = Delta. Tweet 3/8

The Delta Variant was first identified as dominating the start of the third wave in KwaZulu-Natal, here we present recent data, sampled up to 24 June 2021, showing how the expansion of the Delta in KZN is linked to increase in daily cases. 4/8 

The expansion of Delta is also seen in the Western Cape with very recent data produced by @UCT @StellenboschUni and NHLS. 5/8 

We also found that Delta is also causing the expansion in the Eastern Cape (left) and Limpopo (right) and we the Network for Genomics Surveillance is busy expanding sampling around the country. 6/8 



Over 1,500 genomes produced in the last two months and in the last week alone, the Delta genomes from SA deposited in @GISAID grow from 29 to 430, with continuous updates. 7/8
the Network for Genomic Surveillance in South Africa (NGS-SA) needs to thank the very hard work of our laboratory and bioinformatics teams at @krisp_news @StellenboschUni @UCTIDM @nicd_sa @WitsUniversity @UPTuks @UFS and many NHLS laboratories across the country. 8/8
• • •
Missing some Tweet in this thread? You can try to
force a refresh