Evolution of the C.1 lineage in South Africa with multiple Spike mutations - pre-print at: medrxiv.org/content/10.110… - In this tweet thread, I highlight this lineage (assigned as C.1.2), the potential impact of the mutations, and the likelihood that this become a variant of concern
The C.1.2 has 14 mutations in Spike, including three mutations at the Receptor Binding Motif, the Y449H, E484K, and N501Y - two of the mutations have been found in three of the four variants of concern (Alpha, Beta, and Gamma). 

In relationship to decrease neutralization, in addition to the E484K, there are also deletions at position 144 and positions 242-243, the 144 deletions are also seen in the Alpha and the 242-243 in the Beta, which associates with a decrease neutralization of antibodies
There is also the N679K, which is a mutation just upstream of the S1/S2 furin cleavage including Q675H/R, Q677H/P, N679K, and P681H/R have occurred independently in many SARS-CoV-2 global lineages.
In addition, there is also a mutation at 655 positions, the H655Y mutation, which in the 3-D structure of Spike it cluster very close to the S1/S2 furin. Around 8% (i.e. 7/95) of the sequences have also added the P681H found in the Alpha VOC. 

The constellations of mutations worry us as they seem The C.1.2 end up adding multiple mutations that are similar to the Beta with extra mutations at S1/S2 and NTD. Now let me talk more about the C.1 lineages that emerged and persisted in southern Africa.
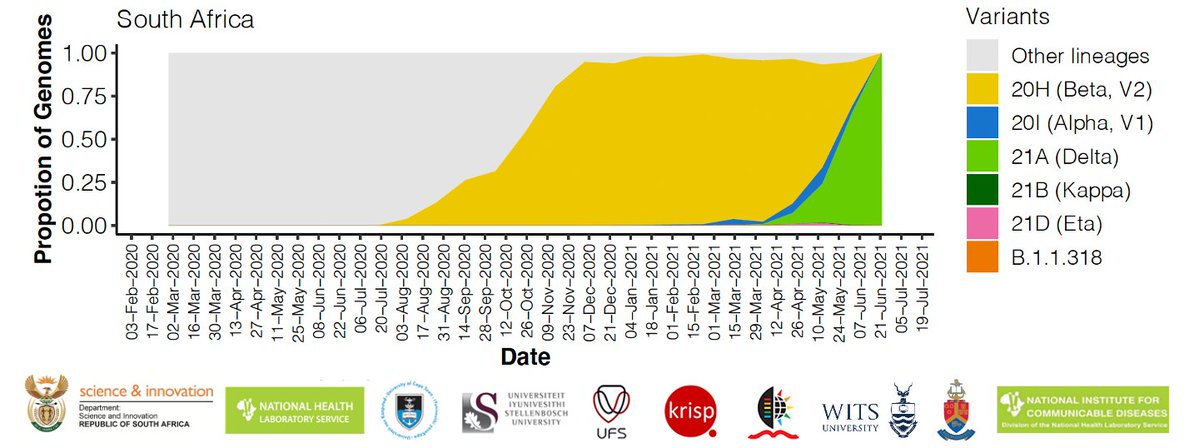
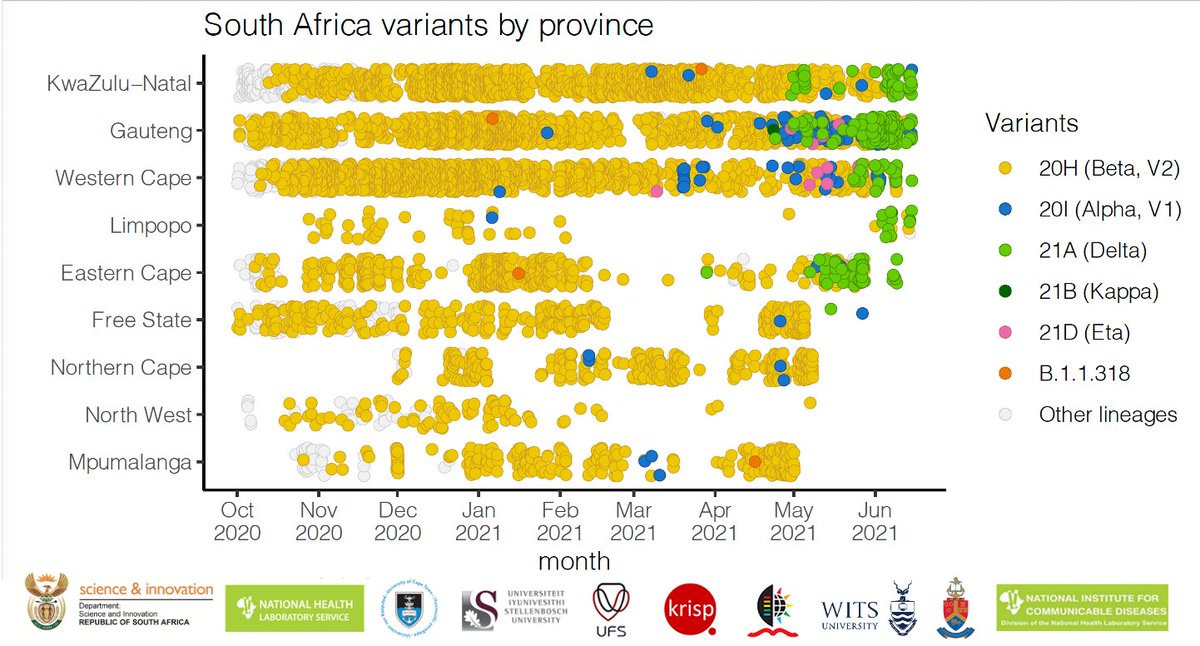
The C.1 was one of the lineages that dominated the first wave of the pandemic in South Africa. The emergence of this lineage is well described in the paper by Tegally et al. Nature Medicine 2021 - nature.com/articles/s4159…
The C.1 emerged in the centre of South Africa (probably Gauteng) and spread quickly to most of the provinces in South Africa during the first wave of infections in 2020. 

The C.1 was also introduced in neighbouring countries and gave rise to a new clade, the C.1.1 in Mozambique, which added the S477N mutation at the RBM and A688S close to the S1/S2. The emergence of the C.1.1. is seen in a pan-Africa analysis pre-print medrxiv.org/content/10.110… 

We now have the C.1.x persisting for over 1 year in South Africa and now evolving to C.1.2 further to add a dozen new mutations at Spike. We are now working with @sigallab & Penny Moore to describe the effect of the mutations in a live virus and pseudo virus system.
The reason why we decided to publish the preprint is that we see the C.1.2 persistence in South Africa and is now in another 10 countries. It is early days as only 95 genomes have been published at GISAID. However, we found that in this pandemic is share info quicker than later.
This is the work of dozens of scientists as part of the Network of Genomic Surveillance in South Africa (NGS-SA). The pre-print analysis was lead by @NICD and @krisp_news and it also involves a very bright and young women scientist, Catherine Scheepers, who is the first author.
Pre-print of C.1.2, which will be updated soon with neutralization results at: medrxiv.org/content/10.110…
• • •
Missing some Tweet in this thread? You can try to
force a refresh