This past week my lab published 4 @biorxivpreprint papers in applied math (biorxiv.org/content/10.110…), biology (biorxiv.org/content/10.110…), bioinformatics (biorxiv.org/content/10.110…), and instrumentation (biorxiv.org/content/10.110…). They were possible thanks to reproducibility... 1/ 






There is a lot of focus on the importance of reproducible science for facilitating replication of published research. That's all good, but reproducible science has another benefit: when adopted by a group it is an incredible accelerant for research *in that group*. 2/ 

Consider the paper we wrote on whole animal multiplexed #scRNAseq. The @GoogleColab notebooks Tara Chari wrote for the analyses were a monumental effort, but she did not start from scratch. 3/
https://twitter.com/lpachter/status/1353731018121859073?s=20
The notebooks were based on two preceding projects in the lab: this paper by @JaseGehring nature.com/articles/s4158…, where the article is accompanied by @GoogleColab for the entire analysis, and biorxiv.org/content/10.110… for which @sinabooeshaghi coded reproducibly. 4/
The bioinformatics paper describing a benchmark of kallisto-bustools and Salmon-Alevin-fry likewise relied on previous reproducible work. @sinabooeshaghi relied on his own notebooks from previous projects, and on reproducible R by @LambdaMoses. 5/
https://twitter.com/lpachter/status/1175067909980246016?s=20
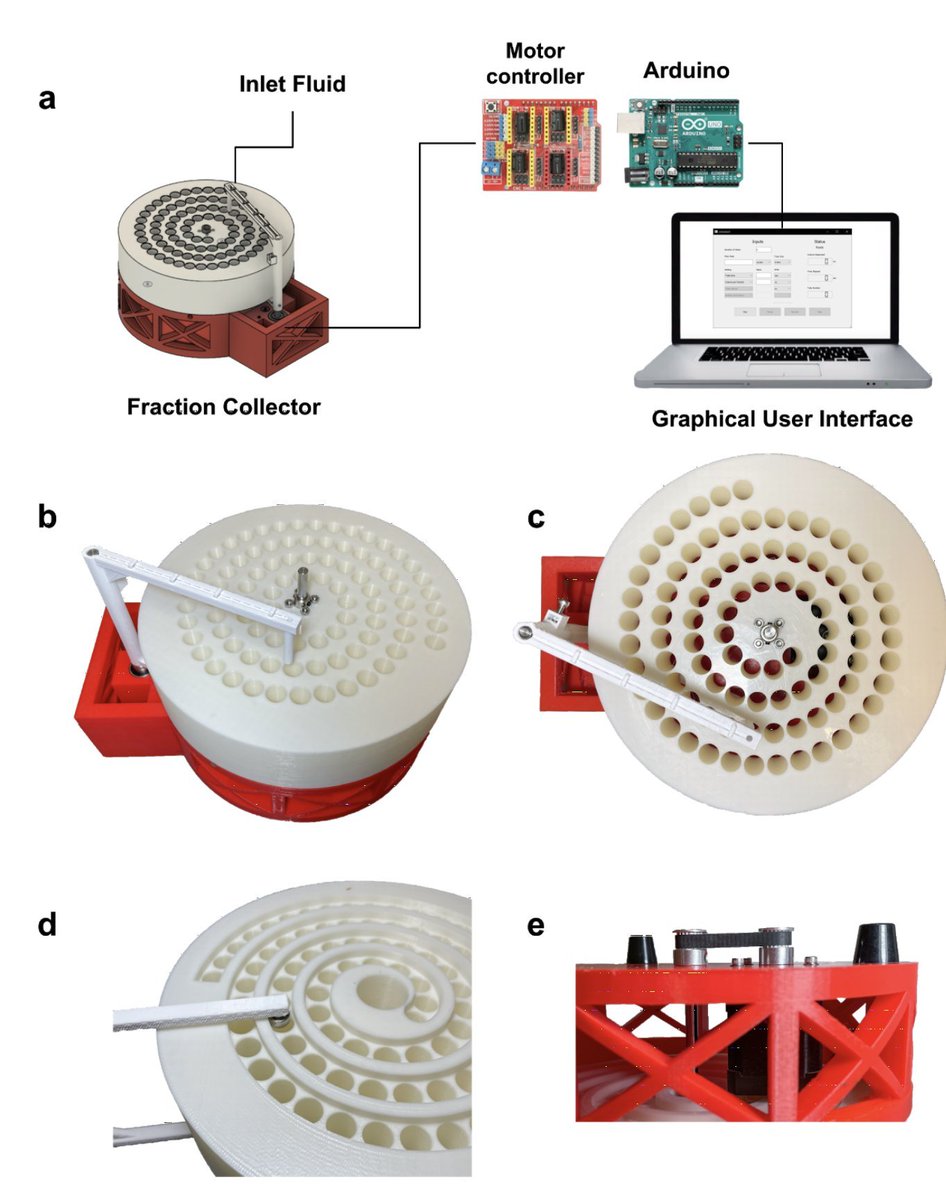
The fraction collector by @sinabooeshaghi, @annekylosaurus et al. was not started from scratch, but with a blueprint from the poseidon project. Making sure poseidon was reproducible in hardware and software was hard work. That work has paid off. 6/
https://twitter.com/lpachter/status/1166380362571767809?s=20
@GorinGennady's work on an exact stochastic simulation algorithm for a transcriptional system benefited from reproducibility standards employed in his previous projects, e.g.
https://twitter.com/lpachter/status/1244794438150316032?s=207/
All of this work benefited from reproducible work by others as well. So while the timing of these preprints was coincidental, their shared foundations are not. Being thoughtful about working reproducibly facilitates progress in science, not just for others, but for oneself. 8/8
• • •
Missing some Tweet in this thread? You can try to
force a refresh