
Opening the Nanopore Community Meeting Tech Update is James Clarke, valiantly speaking through a bad, bad cold. We'll be live tweeting in this thread, with Stu Reid and Rosemary Dokos up next #nanoporeconf
JC: @nanopore accuracy has come a long way. Right now, in just 3-4 seconds, I can pass 1,000 bases of a **single molecule** of DNA through a hole, and know its sequence with over 99% accuracy. That’s better than Q20, by looking at a single molecule - once #nanoporeconf 

JC: Beyond single molecule, you can of course use multiple strands in a pile-up. We’ve achieved Q50 consensus accuracy on bacterial genomes, from 20X coverage. #nanoporeconf 

JC: Beyond single molecule or consensus accuracy, looking to test accuracy – this can be incredibly high on our platform. For examples of high-accuracy and high-output you can look to: @lunadjirackor, @glen_gowers, @methenickname, @decodegenetics, @lw_meredith, @profaxelmeyer 

@lunadjirackor @glen_gowers @methenickname @decodegenetics @lw_meredith @profaxelmeyer JC: we can drive accuracy using: precise nanopore design; engineering of motor enzyme so that it moves the DNA as consistently as possible; and continuous evolution of algorithms to drive better basecalling. We also manage conditions, eg keeping the noise low #nanoporeconf 

@lunadjirackor @glen_gowers @methenickname @decodegenetics @lw_meredith @profaxelmeyer JC: we can drive *output* using nos of nanopores; driven by ASIC design - layer on chemistry - membrane - nanopores; also speed of sequencing (faster - higher output), and a well prepared, well-loaded library to maximise ‘on’ time #nanoporeconf
@lunadjirackor @glen_gowers @methenickname @decodegenetics @lw_meredith @profaxelmeyer JC: our range of flow cells and devices means we can offer the range of outputs needed by users #nanoporeconf nanoporetech.com/products #nanoporeconf
@lunadjirackor @glen_gowers @methenickname @decodegenetics @lw_meredith @profaxelmeyer JC: we have evolved from the R10.3 nanopore to R10.4; as well as accuracy enhancement, this nanopore can be inserted into flow cells in higher numbers. R10.4 is now available on Min, Grid, PromethION #nanoporeconf.
@lunadjirackor @glen_gowers @methenickname @decodegenetics @lw_meredith @profaxelmeyer JC: for a year, PromethION has been capable of delivering a 10Tb run. The device can be tuned for run conditions, with the option to focus on ultra-long reads #nanoporeconf 

@lunadjirackor @glen_gowers @methenickname @decodegenetics @lw_meredith @profaxelmeyer JC: At London Calling, we noted we were developing the PromethION 2 (P2); designed to run up to 2 PromethION flow cells, the idea is to give greater access to low cost, high throughput sequencing, alongside P24 and P48. P2 has built in GPU compute #nanoporeconf 

@lunadjirackor @glen_gowers @methenickname @decodegenetics @lw_meredith @profaxelmeyer JC: also, the PromethION 2 solo, without built in compute, is designed to plug into compute of your choice – eg GridION or your own infrastructure, to perform basecalling and store your data #nanoporeconf 

@lunadjirackor @glen_gowers @methenickname @decodegenetics @lw_meredith @profaxelmeyer JC: Flongle – the smallest flow cell, which connects to an adapter that can be used with the handheld MinION or the desktop GridION. Current limit of the flow cell is 126 channels; but there is still a lot you can do; the record is 2.83 Gb from a flow cell #nanoporeconf
@lunadjirackor @glen_gowers @methenickname @decodegenetics @lw_meredith @profaxelmeyer JC: at London Calling, we noted that we are developing a lower-cost Flongle flow cell, from silicon to lower cost polymer substrate. We have now reached 120 channels on chip design, 2Gb of sequencing data on this. We are moving to a prototype manufacturing phase #nanoporeconf
@lunadjirackor @glen_gowers @methenickname @decodegenetics @lw_meredith @profaxelmeyer JC: we’d like to remove the need for users to flush the flow cell, to make the process easier and more automation-friendly. One route is to use silver instead of platinum; we’re likely to have this in early access on Flongle soon. #nanoporeconf
@lunadjirackor @glen_gowers @methenickname @decodegenetics @lw_meredith @profaxelmeyer JC: the ASIC is the engine of the nanopore flow cell and device. ASICs can consume a lot of power, heat needs to be dissipated through a heat sink. We have been developing a new ASIC, that is low cost and low power, with 400 channels. #nanoporeconf
@lunadjirackor @glen_gowers @methenickname @decodegenetics @lw_meredith @profaxelmeyer JC: our new, low power ASIC has the potential to be used with many types of device, including brand new concepts, and to be powered by eg USB. We have generated sequencing data on the new ASIC, in a breadboard format, and development continues #nanoporeconf 

@lunadjirackor @glen_gowers @methenickname @decodegenetics @lw_meredith @profaxelmeyer JC: even at prototype the new low-power ASIC is delivering respectable accuracy and we expect this to go up as we build dedicated flow cell and instruments #nanoporeconf
@lunadjirackor @glen_gowers @methenickname @decodegenetics @lw_meredith @profaxelmeyer Up next, Stuart Reid, VP Development, outlining how innovation is done at @Nanopore. Process is designed to deliver (a) disruptive innovation (step changes and new approaches), which typically 1st used by developer/early access customers before being more broadly released, and .. 

@lunadjirackor @glen_gowers @methenickname @decodegenetics @lw_meredith @profaxelmeyer ..(b) Continuous improvement. As we learn more about a release.. many of these changes happen seamlessly in the background #nanoporeconf
@lunadjirackor @glen_gowers @methenickname @decodegenetics @lw_meredith @profaxelmeyer SR: I am going to discuss accuracy today, and for that it is important to define what type of accuracy we are talking about, eg raw read simplex, duplex, consensus or test accuracy. #nanoporeconf
@lunadjirackor @glen_gowers @methenickname @decodegenetics @lw_meredith @profaxelmeyer SR: Firstly, raw read – or simplex – accuracy. That’s a single piece of DNA or RNA going through a nanopore, and we are acquiring the signal and basecalling it. We are now beating 99%/Q20 when using the R10.4 nanopore with kit 12 #nanoporeconf 

@lunadjirackor @glen_gowers @methenickname @decodegenetics @lw_meredith @profaxelmeyer SR: we have Duplex accuracies, where we are sequencing both strands of a single molecule and able to reach around Q30 (99.9%). And with @nanopore it is also possible to get information for methylation for free during sequencing #nanoporeconf
@lunadjirackor @glen_gowers @methenickname @decodegenetics @lw_meredith @profaxelmeyer SR: And for consensus accuracy where you have multiple reads and can pile them up, where errors average out with coverage, even modest coverage like 20X can reach beyond Q50 (99.999%) on bacterial genomes when using the latest, nanopore-specific analysis tools #nanoporeconf 

@lunadjirackor @glen_gowers @methenickname @decodegenetics @lw_meredith @profaxelmeyer SR: And of course test accuracy, for accurate insights; in 2020 @nanopore was shown to achieve gold standard sensitivity and specificity on a specific viral detection test #nanoporeconf
@lunadjirackor @glen_gowers @methenickname @decodegenetics @lw_meredith @profaxelmeyer SR: Accuracy improvements have been profound over the years; the ‘R9 pore & kit 10’ was iterated from ~92% to >98% raw read accuracy in ~2 years using algorithm improvement alone. Now we also have the ‘R10.4 pore & kit 12’ for >99% raw read accuracy #nanoporeconf 

@lunadjirackor @glen_gowers @methenickname @decodegenetics @lw_meredith @profaxelmeyer SR: These improvements are delivered through consumables and analysis, with occasional free device upgrades. #nanoporeconf
@lunadjirackor @glen_gowers @methenickname @decodegenetics @lw_meredith @profaxelmeyer SR: now a choice of basecallers, depending on user need. Fast basecallers keep up on all our devices. Or can choose “High accuracy basecaller” (hac) / “super accuracy” (sup) for highest accuracy that is beating Q20/99% raw read accuracy with the latest chemistry. #nanoporeconf 

@lunadjirackor @glen_gowers @methenickname @decodegenetics @lw_meredith @profaxelmeyer SR: in Duplex basecalling, the basecall is made from a pair of nanopore signals (two complementary strands of a single molecule) for higher accuracy. Has been in Bonito, our research basecaller, for a while and now in Guppy so our new kits are duplex enabled but still in beta.
@lunadjirackor @glen_gowers @methenickname @decodegenetics @lw_meredith @profaxelmeyer SR: SR: for consensus accuracy we recommend our own Medaka tool, although the output from the assembler (Flye) is also good. Best results with R10.4 and kit 12 (eg >Q50 bacterial) and R9+kit10 also produce good results. #nanoporeconf
@lunadjirackor @glen_gowers @methenickname @decodegenetics @lw_meredith @profaxelmeyer SR: onto variant calling, we are changing our recommendation for SNPs and indels to Clair3, developed by Ruibang Lo @acquaskyline and team
(replacing Medaka, which we still recommend for consensus). Other methods include PEPPER-Margin-DeepVariant #nanoporeconf
(replacing Medaka, which we still recommend for consensus). Other methods include PEPPER-Margin-DeepVariant #nanoporeconf
@lunadjirackor @glen_gowers @methenickname @decodegenetics @lw_meredith @profaxelmeyer SR: with our tool recommendations, on 60X native data, this is the variant calling – variants include methylation - you should be achieving across the whole genome #nanoporeconf 

SR: in summary, here are the variant calling stats with the specifically recommended tools #nanoporeconf 

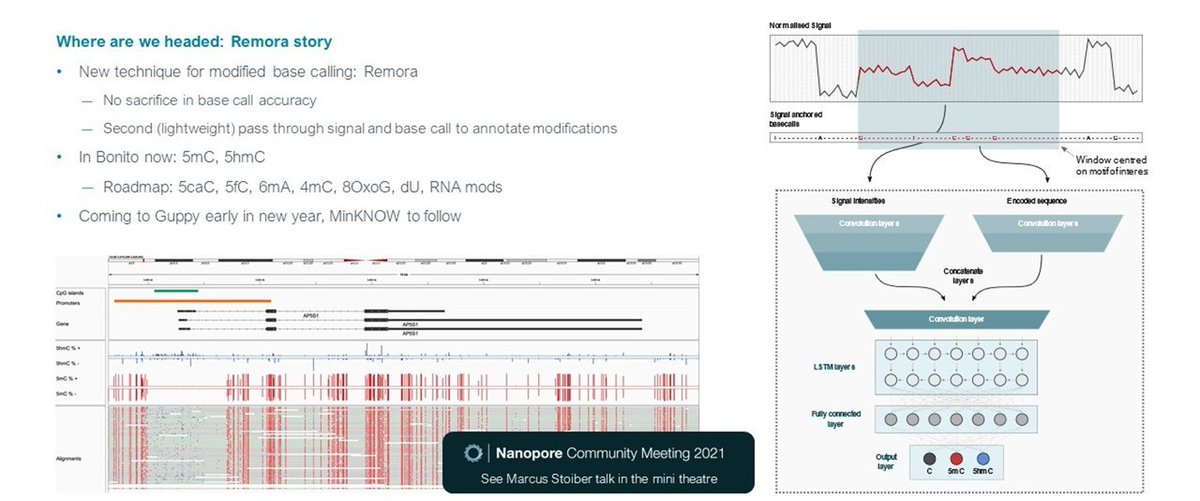
SR: We have a new method for methylation – Remora - with which we wish to set a new gold standard. We believe R9.4.1 was already better than bisulfite, and R10.4 is even better #nanoporeconf 

SR: Remora is 2nd, lightweight pass through the signal, that can run with basecalling, sacrificing no accuracy when delivering methylation analysis in the same experiment. Now in Bonito for 5mC and 5hmC - roadmap for broader DNA & RNA mods and into Guppy in new yr #nanoporeconf 
SR: if you would like to test these results, you can either buy a MinION and try yourself, or look at our open datasets. In EPI2ME Labs there are many workflows and tutorials that can help you reproduce those results
SR: barcoding on nanopore has been accurate for many years. You can choose single-ended barcodes, or if you use both ends you can reach 99.99% correct barcode assignation #nanoporeconf 

SR: where we see lower accuracy than expected, we like to look at the sample to understand more. In this plant example, PCRing the sample closed the accuracy gap-we believe base mods were causing issues. Basecaller has now been trained to “flatten” those mods. #nanoporeconf 

SR: users may wish to detect those mods directly, however we now want to basecall canonical bases , and characterise modifications with Remora. Recommend @aw_ngs talk on plant genomes in the mini theatre
#nanoporeconf
#nanoporeconf
@AW_NGS SR: @nanopore can, and always has been able to, sequence any length fragments, including short fragments #nanoporeconf 

SR: Short Fragment Mode (SFM): In our software config, we have had a lower limit on the length of fragments that are written out. SFM=> write out fragments down to ~20 bases. Targeting release of this SFM in MinKNOW in the new year, but contact support 4 access now #nanoporeconf 



SR: in summary here are the accuracy levels that can be achieved with the latest and most appropriate chemistry and tools #nanoporeconf 

SR: some updates on our software. As you know MinKNOW is the instrument software. It is designed to be point-and-click, but supports nanopore versatility with expert features below the surface #nanoporeconf 

SR: MinKNOW is aiming to maximise your output, maximise your accuracy, and minimise your time to answer. We are planning a range of additional improvements too #nanoporeconf 

SR: Basecalling continues to get faster, and we will support a wider range of platforms and hardware with auto-configuration features. #nanoporeconf
SR: Adaptive sampling-unique to Oxford @nanopore, where you can ‘taste’ DNA in real time and ‘spit it out’ of the nanopore in favour of the next strand, if not of interest. We've integrated a few of the simple features eg target enrichment and panels. Here: 200-gene cancer panel 

SR: Adaptive sampling has an API that third party tools can be developed on, please join Sam Kovaka and Alex Payne in the Adaptive sampling breakout session #nanporeconf @samkovaka @alexomics
@samkovaka @alexomics Up next, Rosemary Sinclair Dokos, VP, Product. RD reminds us of the features of nanopore sequencing #nanoporeconf 

@samkovaka @alexomics RD: we are making some changes to our store offering, adding a 12 – pack of flow cells for Flongle, and expanding the pricing choices for PromethION around the lower number of flow cells, to prepare for more P2 users #nanoporeconf
@samkovaka @alexomics RD: Cost per Gb of nanopore data is competitive across a range of scenarios, whether performing high-throughput human genomes w PromethION for as little as $600 per flow cell, driving as little as $3 a Gb at 200Gb per flow cell, or a $67.5 Flongle flow cell for a rapid analysis 

@samkovaka @alexomics RD: the platform is very versatile, and users can choose the best fit for their experiment; the answer to “how much data will I get” often depends on which levers that you vary #nanoporeconf 

@samkovaka @alexomics RD: the rich biology delivered by nanopore sequencing is being used to untangle complex biology, where the genetic cause of disease may be a combination of multiple types of genetic variation, that may also interact #nanoporeconf 

@samkovaka @alexomics RD: we apply the same principles to product release as to innovation; initial engagement with developers and early access users evolves into fully released products, allowing us to move fast and continuously improve #nanoporeconf 

@samkovaka @alexomics RD: this is what continuous iteration looks like; ongoing improvement delivered through consumables and software #nanoporeconf 

@samkovaka @alexomics RD: we have complex, cross-platform upgrades to deliver to you. We will continue to support people using the established R9 nanopore and kit 10, and are incl multiple upgrades in that kit. But we are also releasing Kit 12, which includes the recent Q20+ chemistry #nanoporeconf 

@samkovaka @alexomics RD: we have a range of preparation kits to support almost any experiment. And now, we can support inputs of 50-100ng of DNA in the rapid/field kit, down from 400ng; we are working on pushing this improvement across the whole range. #nanoporeconf
@samkovaka @alexomics RD; we’re launching today the native barcoding kit for R10.4 & kit 12, so that the Q20+ chemistry can be multiplexed. 96 barcodes enables very competitive cost per sample. We are also releasing the rapid kit for Q20+ chemistry #nanoporeconf
@samkovaka @alexomics RD: we are releasing the multiplex ligation kit to enable two human genomes per PromethION flow cell; this in combination with automation-the Hamilton script for 2 genomes per FC is ready – come and talk to us about it! #nanoporeconf 

@samkovaka @alexomics RD: to support ultra-long read sequencing, we recently launched the NEB Monarch kit Works with protocols for cells/blood, and soon, tissue. Our apps team is happy to support conversations about extraction methods in the community #nanoporeconf 

@samkovaka @alexomics RD – using Adaptive Sampling for reduced representation methylation sequencing is now available in MinKNOW. 6 million CpG islands called on a MinION; we think that’s a superior performance to bisulfite #nanoporeconf 

@samkovaka @alexomics RD: the cDNA PCR sequencing kit is live in store today - it can generate >160M 1kb reads from a PromethION flow cell, and >20M reads from a MinION flow cell; this kit includes fuel fix adapter for more hands off time, accurate Poly(A) tail estimate, UMIs #nanoporeconf
RD: the cDNA PCR kit also enables single cell, delivering full length reads for isoform discovery/characterisation, gene fusions, cell subtype, spatial. Emerging work in the community has developed enrichment strategies; analysis pipeline in development #nanoporeconf 

RD: By partnering with PCS, we can improve the no of reads you can get off a flow cell; this protocol is now in the community and we would love to hear from users so we can continue to develop this single cell offering #nanoporeconf
RD: in the summer we released the Midnight kit for SARS-CoV-2 sequencing, based on @nikki_freed method of rapid, low cost COVID sequencing that performs well even at high CT values (from $9.55 per sample) #nanoporeconf nanoporetech.com/covid-19
@Nikki_Freed pls see update earlier on Midnight and #omicron #SARSCoV2
https://twitter.com/nanopore/status/1466072171328032775?s=20
@Nikki_Freed RD: short fragment mode, as previously summarised by SR, is now available for developers so that users can sequence high volumes of short fragments, but retain methylation, portability, ease of use and accessibility #nanoporeconf
@Nikki_Freed RD: no sample prep talk is complete without VolTRAX, which now includes PCR-enabled devices that are on their way to community users #nanoporeconf
@Nikki_Freed RD: thinking about MinION vs GridION, you can choose the most appropriate for your sample needs #nanoporeconf 

@Nikki_Freed RD: which PromethION device should you choose? For ~200 PromethION flow cells a year, the P2. For ~2,000 a year, P24. And for more than 4k-4.5k flow cells, P48 #nanoporeconf 

RD: you can now pre-order the P2 solo on the store. At $10,455 for a starter pack we believe this puts sub-$1000, comprehensive human genomes in the hand of any scientist #nanoporeconf nanoporetech.com/products/p2 

RD: the dream is to load the dataset, see the files you want to see come out at the end. Teams driving towards sample in and BAM VCF/fastq out, with the right analyses in between #nanoporeconf
RD: we offer a range of analysis options, from research software that is at the cutting edge, to end to end EPI2ME workflows #nanoporeconf 

@threadreaderapp unroll
• • •
Missing some Tweet in this thread? You can try to
force a refresh







