UK reports Delta Omicron recombination, this time it's the real thing.
I will try to answer 3 questions here:
1.Was this expected?
2.Is this the report from Cyprus?
3.Should we be concerned?
1/8
I will try to answer 3 questions here:
1.Was this expected?
2.Is this the report from Cyprus?
3.Should we be concerned?
1/8
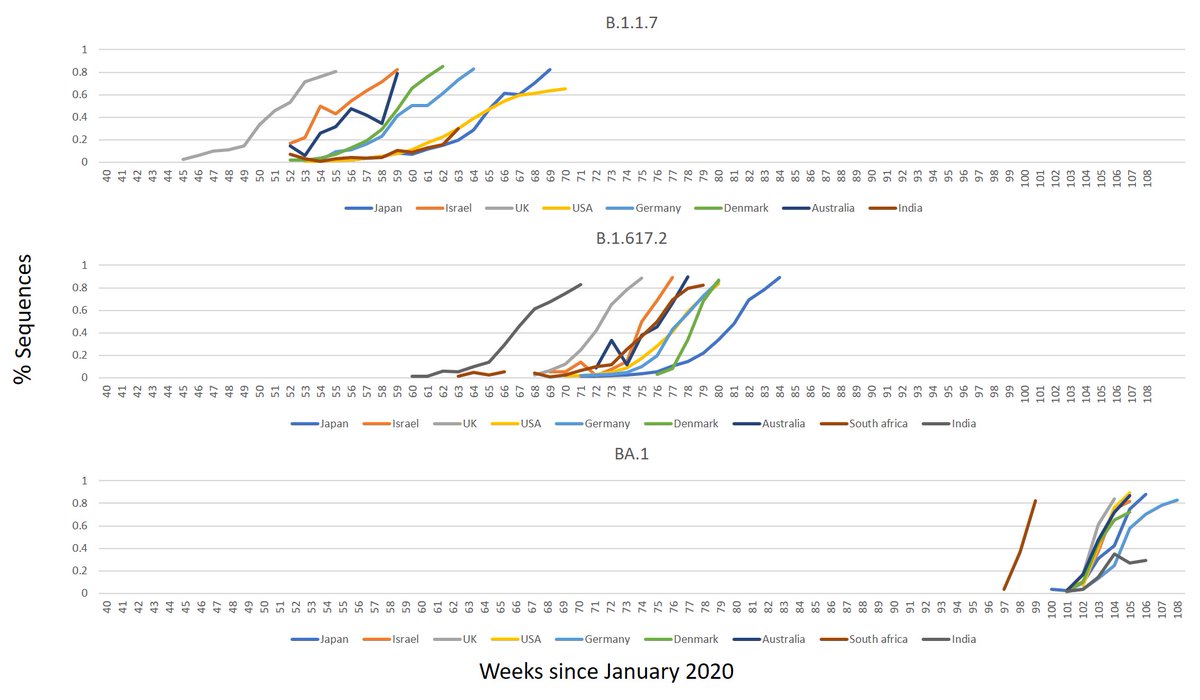
1.Was this expected? Yes.
Recombination occurs when a cell is infected with 2 variants.
when 2 variants are in high # & % recombination is more likely to be visible. it is likely to occurred during the Δ times, but if AY.4 recombined with AY.3, that's gonna be hard to spot
2/8
Recombination occurs when a cell is infected with 2 variants.
when 2 variants are in high # & % recombination is more likely to be visible. it is likely to occurred during the Δ times, but if AY.4 recombined with AY.3, that's gonna be hard to spot
2/8
For example:
When α rose and mostly replaced B.1.177 in the UK, XA (recombinant of those 2) detected.
github.com/cov-lineages/p…
When Delta rose and replaced α, XC detected.
github.com/cov-lineages/p…
And now – UK spotted some Δ-ο recombination.
gov.uk/government/pub…
3/8
When α rose and mostly replaced B.1.177 in the UK, XA (recombinant of those 2) detected.
github.com/cov-lineages/p…
When Delta rose and replaced α, XC detected.
github.com/cov-lineages/p…
And now – UK spotted some Δ-ο recombination.
gov.uk/government/pub…
3/8
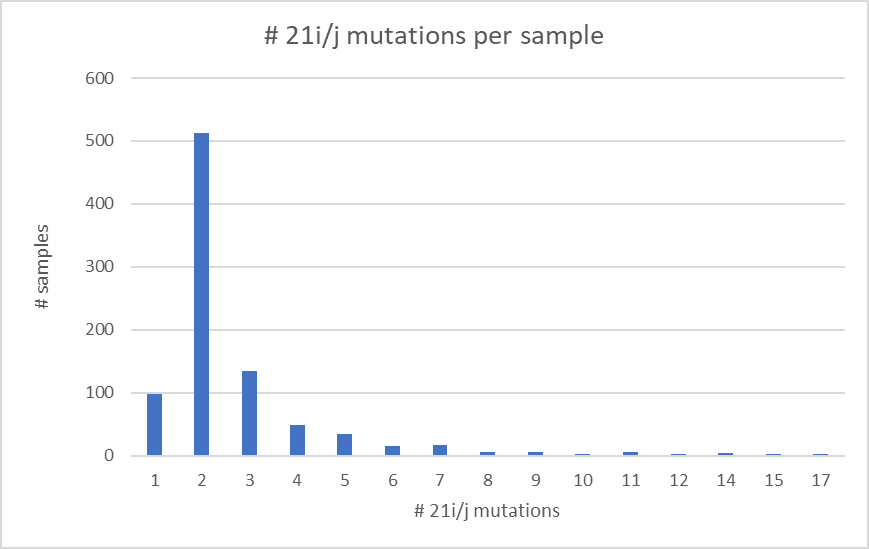
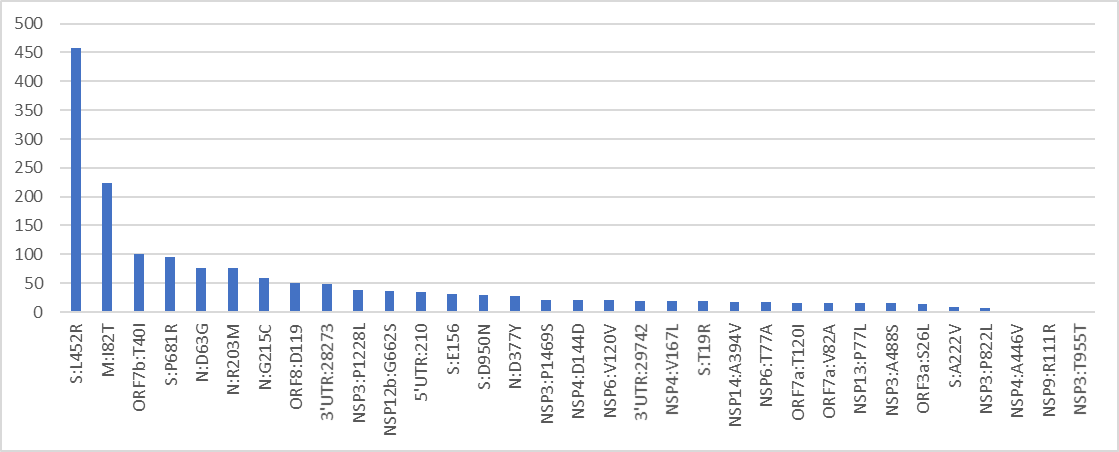
With an algorithm developed by @DrorBendet I found additional Δ-ο suspects .
Among these are the lineage shown here which contains 2 breakpoints (2 parts of Δ with one part O in the middle).
nextstrain.org/fetch/genome.u…
4/8
Among these are the lineage shown here which contains 2 breakpoints (2 parts of Δ with one part O in the middle).
nextstrain.org/fetch/genome.u…
4/8
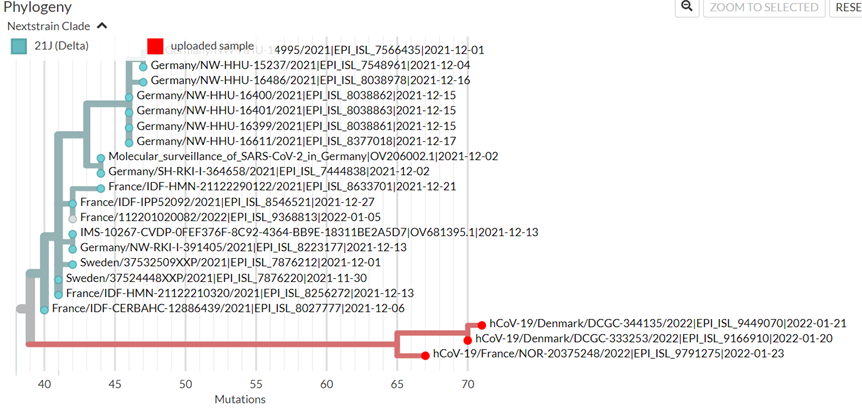
2. Is this the report from Cyprus? No!
Those were just Δ sequences (Complete Δ, no breakpoint), from different AY lineages, that had ~2-3 mutations that also appeared in o.
Δ sequences with additional mutations like these found long before o emerged.
5/8
Those were just Δ sequences (Complete Δ, no breakpoint), from different AY lineages, that had ~2-3 mutations that also appeared in o.
Δ sequences with additional mutations like these found long before o emerged.
5/8
Should we be concerned? Hard to say (but probably not).
Recombination is just another tool in the evolutionary toolbox of the virus. Like mutations that accumulate in a chronic infection, reverse zoonosis or even genetic drift from transmission in the community.
6/8
Recombination is just another tool in the evolutionary toolbox of the virus. Like mutations that accumulate in a chronic infection, reverse zoonosis or even genetic drift from transmission in the community.
6/8
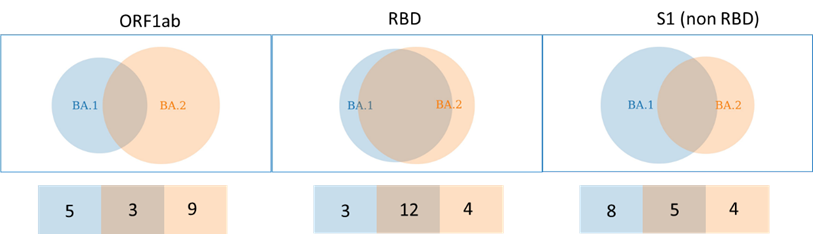
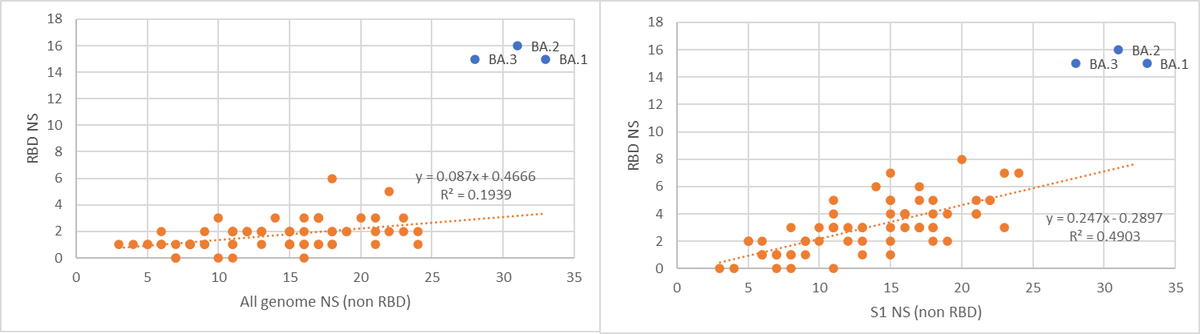
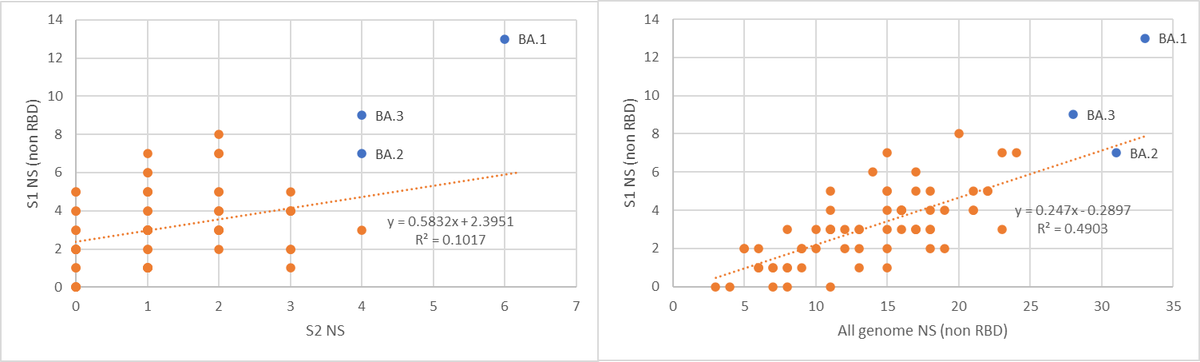
Recombination is not the combination of mutations from both parents. Each variant contributes a part, but the complementary part disappears. For example, the unsuccessful BA.3 is a recombinant of BA.1 and BA.2 (which are causing the main part of the pandemic right now).
7/8
7/8
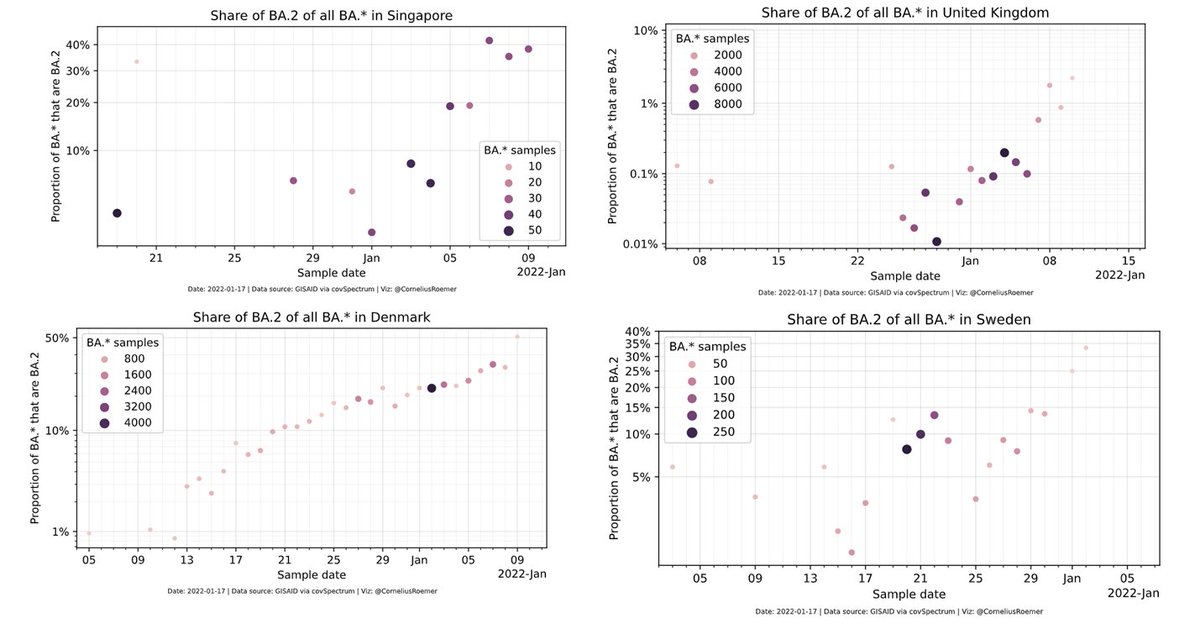
Remember - The cases of α - B.1.177 ,Δ-α and now ο-Δ found in countries with high levels of 🧬. That may be attributed to those being really rare phenomena.
Successful Variants defined as VOC / VUI / VUM originated a lot from places with low 🧬 such as SA, India and Peru.
8/8
Successful Variants defined as VOC / VUI / VUM originated a lot from places with low 🧬 such as SA, India and Peru.
8/8
• • •
Missing some Tweet in this thread? You can try to
force a refresh