
We made the cover! Presenting the #photosystem of Gemmatimonas phototrophica, a funky little bacterial alga (the first discovered example of its genus) found in the Gobi desert a little under a decade ago. This one's been quite the journey, so buckle in. (1/) 

What I'd mostly like to talk about is the saga of how this model came to be... but first, a little about what makes it special. At first glance, it's fairly similar to other bacterial photosystems: a central reaction complex (RC) surrounded by a light-harvesting (LH) ring. (2/)
What's brand new here is the way it increases its light-harvesting capacity. All other known photosynthetic bacteria have a single LH ring around the RC (termed LH1), and then make smaller "satellite" rings (LH2) which hang around the edges. (3/)
Gemmatimonas took its own path. After acquiring the core photosynthetic genes from another bacterium some time in the Precambrian, it found a way to assemble an entire extra LH ring around the core complex. (3/)
Doing this efficiently is a bigger biological challenge than you might first think, because what these complexes actually *do* with the light energy is to reduce quinones - rather large membrane-resident molecules that have to find their way in and out of the complex. (4/)
Here's one shown in green spacefill... that thing has to worm its way through this seemingly-impenetrable wall (in both directions) to complete the catalytic cycle. How does it do that? Answers later. First, a bit on how this came to be. (5/) 

This is actually the 5th photosynthetic complex I've published with @PuQian3 and Neil Hunter, but as these things often go it's actually the first one we worked together on. Pu first approached me back in the pre-pandemic times of mid-2019 (seems much longer, doesn't it?) (6/)
He and Neil had been working with a group led by Michal Koblizek and Alastair Gardiner at #algatech #centrumalgatechtrebon, and had a mid-3Å map that they were finding challenging to model. One look and I was hooked. (7/)
That original map turned out to be a bit weird. Knowing what I know now I'd probably recognise the issue pretty quickly, but at the time I didn't know what was going on. The double-ring system was clear and easy enough to model, but the RC was *really* messy. (8/)
After spending a lot of time struggling with it, I eventually noticed some ghostly density for a couple of extra, unexpected transmembrane helices - either very mobile, or at partial occupancy. After much discussion, the team decided to collect a new #cryoEM dataset. (9/)
To do this, they enlisted the help of @cjrlab and Katerina Naydenova, with their fancy new movement-minimising grid design (science.org/doi/full/10.11…). When @PuQian3 got back to me with four(!) new maps, the difference was nothing short of spectacular. (10/)
So, what had gone wrong with the original? Well, it turns out that there were actually two alternative conformations of the outer (LHh) ring (packed as 3 LHh subunits per 2 LH1), present in roughly-equal proportions (see the excellent supplementary figure from @cjrlab). (11/) 

Turns out if you consider only the protein components of the rings, then a 22.5 degree rotation leads to a near-perfect overlay... which is what the original reconstruction had done, giving a decent ring map at the expense of the RC. (12/)
The "extra" helices I'd noticed were simply the resulting artifacts. Anyway, that alternate conformation explains two maps. The other two arise from the presence or absence of the extra cytoplasmic "RC-U" chain (another Gemmatimonas novelty). (13/)
The other new chain the map turned up was the periplasmic "RC-S" (the red chain in the side and top views here). As unexpected novelties, RC-S and RC-U had to be identified by sequencing from the map, which was surprisingly easy (did I mention these maps were *gorgeous*?) (14/) 

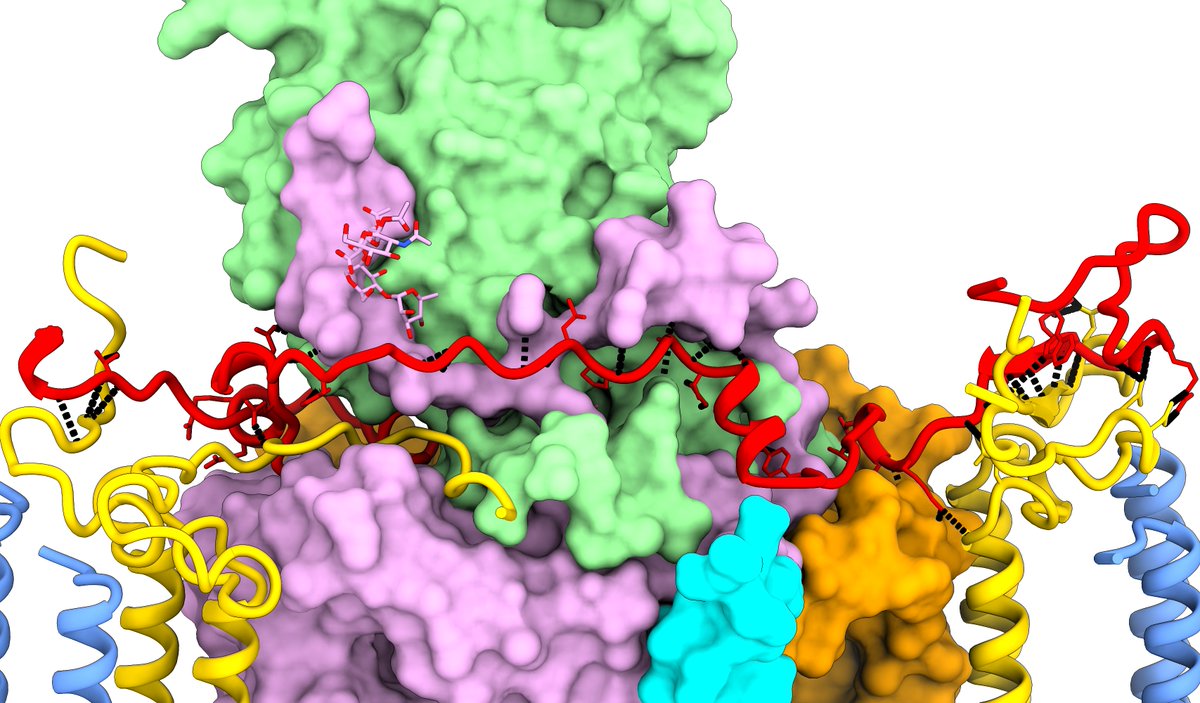

Now, I must admit I'm occasionally prone to flights of fancy. When I first saw these two alternate ring states and the RC-S looking for all the world like the hands of a clock, I desperately wanted there to be some sort of "stepper motor" like mechanism going on. (15/)
Once I started seriously modelling the ligands that idea was quickly laid to rest. The "back" face of the RC (away from the quinone cavity) contains a cohort of tightly-packed lipids bound to the RC, with legs protruding into the LH1 ring preventing any rotation. (16/) 

Still, it was a fun idea while it lasted. Moving on... (16/)
A little tangential to the core biology, but a real pleasure (particularly for the #glycotime folks) was the unexpected discovery a pair of super-clear O-linked glycans on the RC (as far as I know, a first for bacterial photosystems?) (17/)
Unfortunately our resources didn't extend to identifying these by biochemical means (a huge job in its own right), but with a little help from @glycojones sanity-checking the G. phototrophica genome for relevant enzymes we came up with a pretty solid assignment. (18/)
This includes the first instance in the PDB of a 2,3-di-O-acetyl glucuronic acid (a known bacterial sugar modification, just never resolved in a structure before). (19/)
Anyway, back to the mystery of how the quinones get in and out. Initially that seems impossible - when everything's modelled the ring seems like an impermeable wall. But ultimately I think it comes down to the outermost, most weakly-bound ring of bacteriochlorophylls... (20/)
These were actually another unexpected discovery - their density was too weak/blurry to assign in the original map. We modelled the complex with all of these present (and there's definitely at least some density at each site). But... (21/)
... once you look at an overview, there's a really clear strong-strong-weak pattern for these guys, where every third one seems only partially occupied. Notably, this is most pronounced around the quinone cavity. (22/)
(Quinone cavity is on the right, modelled quinones in green). Take one of these chlorophylls away, and you get a tunnel you could drive a truck through. (23/) 

Anyway, I should wrap it up there. This was of course a huge team effort by some really great people. One final special shout-out to @laurencepearl who taught me how to do Fresnel glow effects in Blender - the cover image was actually my first serious Blender project. (fin)
• • •
Missing some Tweet in this thread? You can try to
force a refresh




