
Four new preprints were posted in the last few days about the #Huanan #market and #COVID19 #Origin
1/
1/

Some scientists tend to think that an initial animal-to-human transmission occured at this market (2 new preprints going in that direction)
whereas others believe that the market was simply the location of an early ‘superspreading’ event (the new CCDC preprint).
2/
whereas others believe that the market was simply the location of an early ‘superspreading’ event (the new CCDC preprint).
2/
The four new preprints do not bring definitive, conclusive evidence on this issue. It remains unclear how patient zero at the Huanan market was contaminated and whether the market was a site of animal-to-human contamination.
3/
3/
The Chinese CDC preprint: extensive sampling study,
no animal sample was tested positive.
(see detailed comments on the web page ⤵️)
researchsquare.com/article/rs-137…
4/
no animal sample was tested positive.
(see detailed comments on the web page ⤵️)
researchsquare.com/article/rs-137…
4/
No mention of racoon dogs in this preprint, although a previous study reported their presence in the market in November-Dec 2019.
nature.com/articles/s4159…
➡️ Were racoon dogs absent when the Chinese CDC team sampled at the market?
5/
nature.com/articles/s4159…
➡️ Were racoon dogs absent when the Chinese CDC team sampled at the market?
5/
Worobey et al preprint : the market was the epicenter of SARS-CoV-2 emergence and early COVID-19 disease transmission.
zenodo.org/record/6299600…
➡️ Indeed. This conclusion is consistent with both a zoonotic and lab-related origin.
6/
zenodo.org/record/6299600…
➡️ Indeed. This conclusion is consistent with both a zoonotic and lab-related origin.
6/
Worobey et al. shows that the market was the site of an early superspreading event.
At the begining of COVID outbreak, there was:
-low trasmission rate
-high variation in transmission rate (kappa)
so many chains of transmission died out and
7/
At the begining of COVID outbreak, there was:
-low trasmission rate
-high variation in transmission rate (kappa)
so many chains of transmission died out and
7/
as a result, an early superspreading event can look a posteriori like the epicenter of the outbreak.
Even more so with the asymptomatic cases which were never detected.
8/
Even more so with the asymptomatic cases which were never detected.
8/
A lab manipulating coronaviruses is located at 8 min walk from the market. So Worobey et al. analysis also shows that this lab is the epicenter of SARS-CoV-2 emergence!
Frankly, the stakes are too high to draw a strong conclusion from Worobey et al. statistical analysis.
9/
Frankly, the stakes are too high to draw a strong conclusion from Worobey et al. statistical analysis.
9/
Pekar et al preprint: a model with 2 zoonotic events is consistent with the data.
zenodo.org/record/6291628…
➡️ Lineages A & B differ by only 2 mutations & very few sequences are available from the early days of the pandemic. It is therefore not possible with available data..
10/
zenodo.org/record/6291628…
➡️ Lineages A & B differ by only 2 mutations & very few sequences are available from the early days of the pandemic. It is therefore not possible with available data..
10/
to fully exclude an alternative hypothesis, proposed by Kumar et al in 2021 (academic.oup.com/mbe/article/38…), that there was a single zoonotic event a few weeks before December 2019.
11/
11/
WHO-China joint report mentions 100 laboratory-confirmed cases in Dec 2019, ie patients from which SARS-CoV-2 RNA was extracted. Yet only ~20 SARS-CoV-2 genome sequences from Dec 2019 are available.
Seq data from the other patients is highly needed.
who.int/news/item/30-0…
12/
Seq data from the other patients is highly needed.
who.int/news/item/30-0…
12/
And last Huanan market preprint by @franciscodeasis & myself: we propose that several early infections at the Huanan market probably occurred via human-to-human transmission in closed spaces such as canteens, Mahjong rooms or toilets.
zenodo.org/record/6300876…
13/
zenodo.org/record/6300876…
13/
The earliest detected cases are vendors that are too far away from each other to be contaminated by virus spreading from one (or two) animal sources.
14/
14/
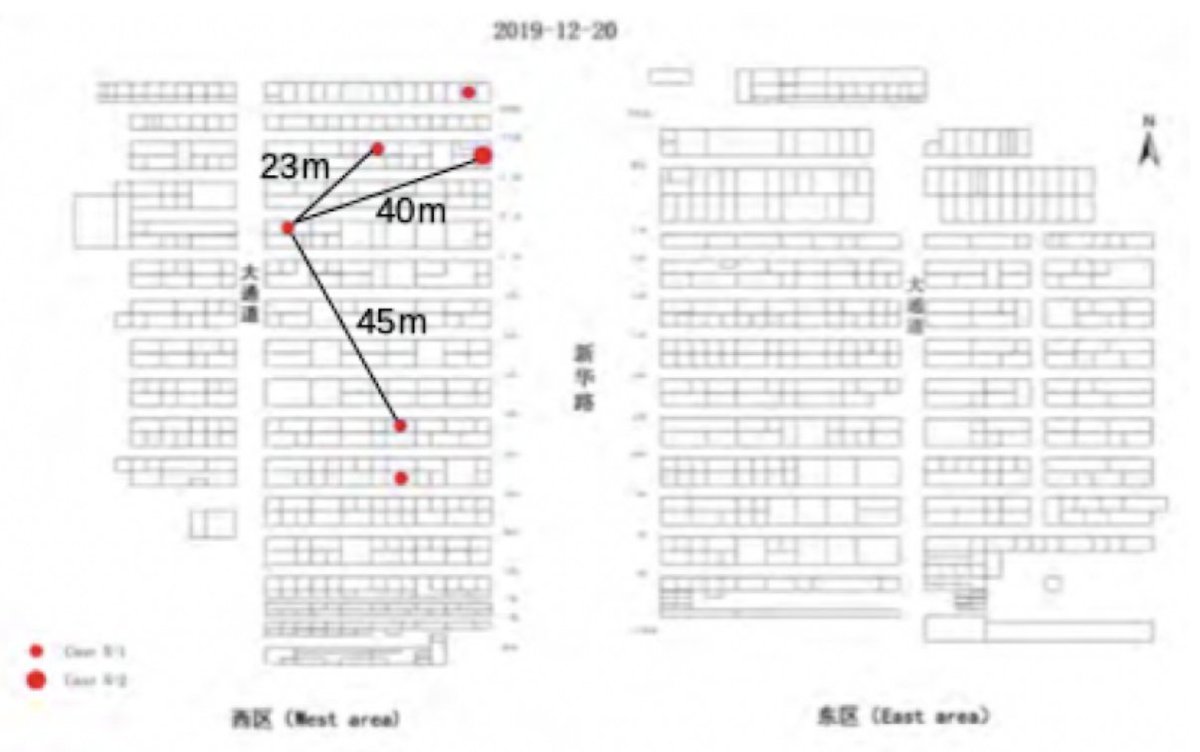
Early market patients may have had contact with each other in the toilets, canteens, or Mahjong rooms. More epidemiological studies of the market patients are needed.
15/
15/
Even if several early market outbreak cases may turn out to be explained by human-to-human transmission, it remains unclear how the first person at the Huanan market was contaminated and whether the market was a site of animal-to-human contamination.
end/
end/
• • •
Missing some Tweet in this thread? You can try to
force a refresh







