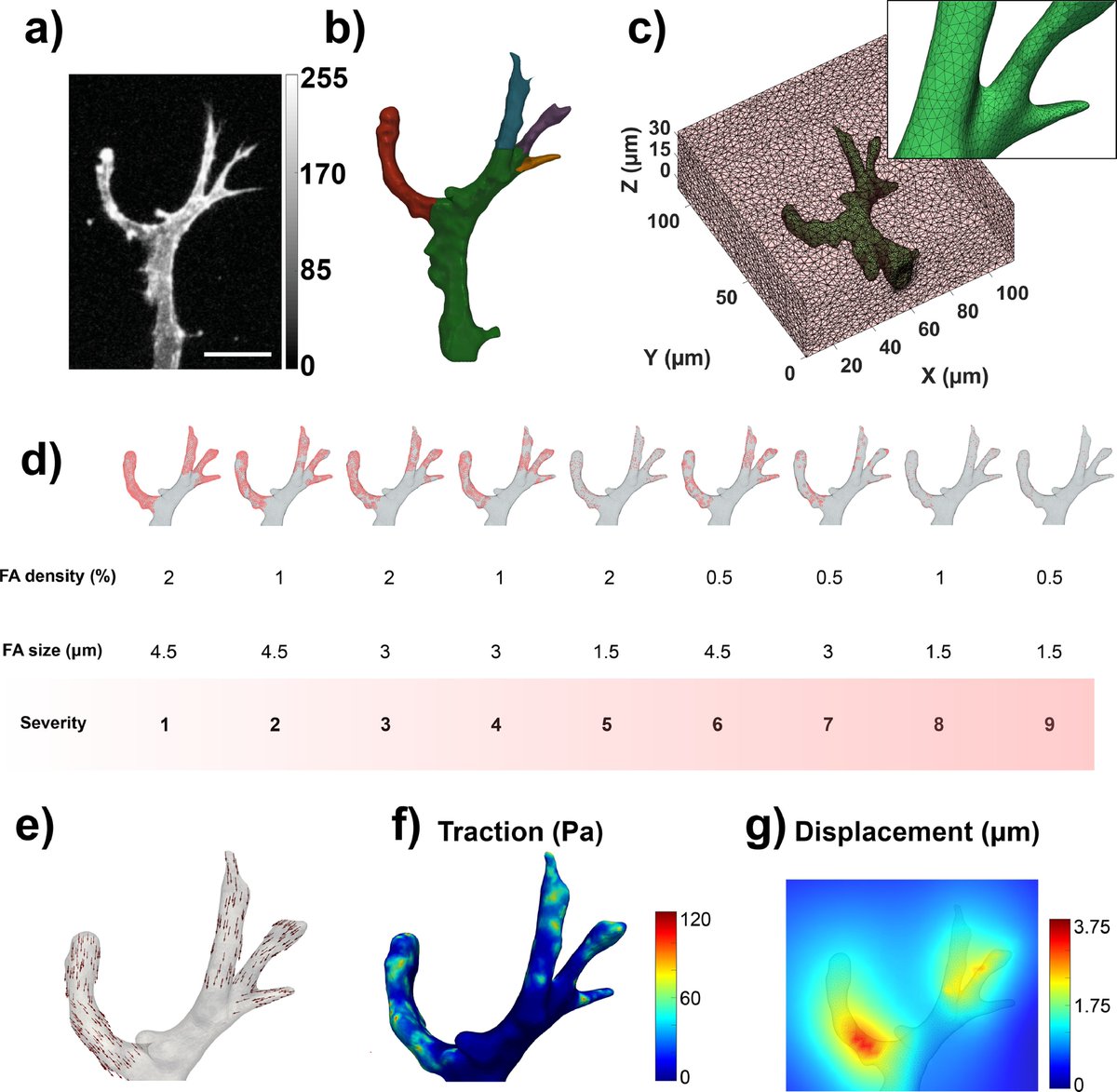
Last April I defended my #PhD at @KU_Leuven, in which I developed ways of looking at the #Force within your cells...
(1/20) 🧵In this #ThesisThread I will tell you exactly how I did it...
(1/20) 🧵In this #ThesisThread I will tell you exactly how I did it...
(2/20) No, at least in our galaxy, our cells do not have #midichlorians. But all ~30 trillion cells of your body are able to:
🔍Change their behavior if they #feel changes in the forces around them...
💪Exert #forces to move or to explore the environment
🔍Change their behavior if they #feel changes in the forces around them...
💪Exert #forces to move or to explore the environment
(3/20) Let me give you an example:
🚀When astronauts spend time in space, their bones become weaker! This means that the cells of their bones take a decision when they don't feel the #gravity anymore!
If there is no need, why should cells bother and maintain bones?
🚀When astronauts spend time in space, their bones become weaker! This means that the cells of their bones take a decision when they don't feel the #gravity anymore!
If there is no need, why should cells bother and maintain bones?

(4/20) Another example: many cells in our body need to move from point A to point B.
👀 Look at this beautiful video of a cell migrating and pulling on collagen fibers (in green) to push itself forward!
(collagen is present everywhere in your body! for ex. in your ear!👂)
👀 Look at this beautiful video of a cell migrating and pulling on collagen fibers (in green) to push itself forward!
(collagen is present everywhere in your body! for ex. in your ear!👂)
(5/20) Ok! So far we've learned that cell forces exist, sure... But why do we care about them?🤔
It turns out that these forces are also very important in #cancer...
👀Look at how these cancer cells (in green) pull on collagen (in red) to invade tissues outside the tumor!!
It turns out that these forces are also very important in #cancer...
👀Look at how these cancer cells (in green) pull on collagen (in red) to invade tissues outside the tumor!!
(6/20) You may be realizing now that researchers are actually very interested in understanding how cells generate such forces👨🔬👩🔬.
If we know how a cancer cell exerts force to invade other tissues, why not try to stop them?
So the next question is how do we study cell forces?
If we know how a cancer cell exerts force to invade other tissues, why not try to stop them?
So the next question is how do we study cell forces?
(7/20) Since studying tissues with millions of cells is very complicated, researchers typically use #invitro cell cultures. This means that we culture them on a dish.
This way, we can have more control of the situation and do simple experiments that give us useful information.
This way, we can have more control of the situation and do simple experiments that give us useful information.

(8/20) You may be wondering now... ok, I have some cells in the dish 🧫. How on Earth can I see the #forces that they exert!!?
Let me give you an example.
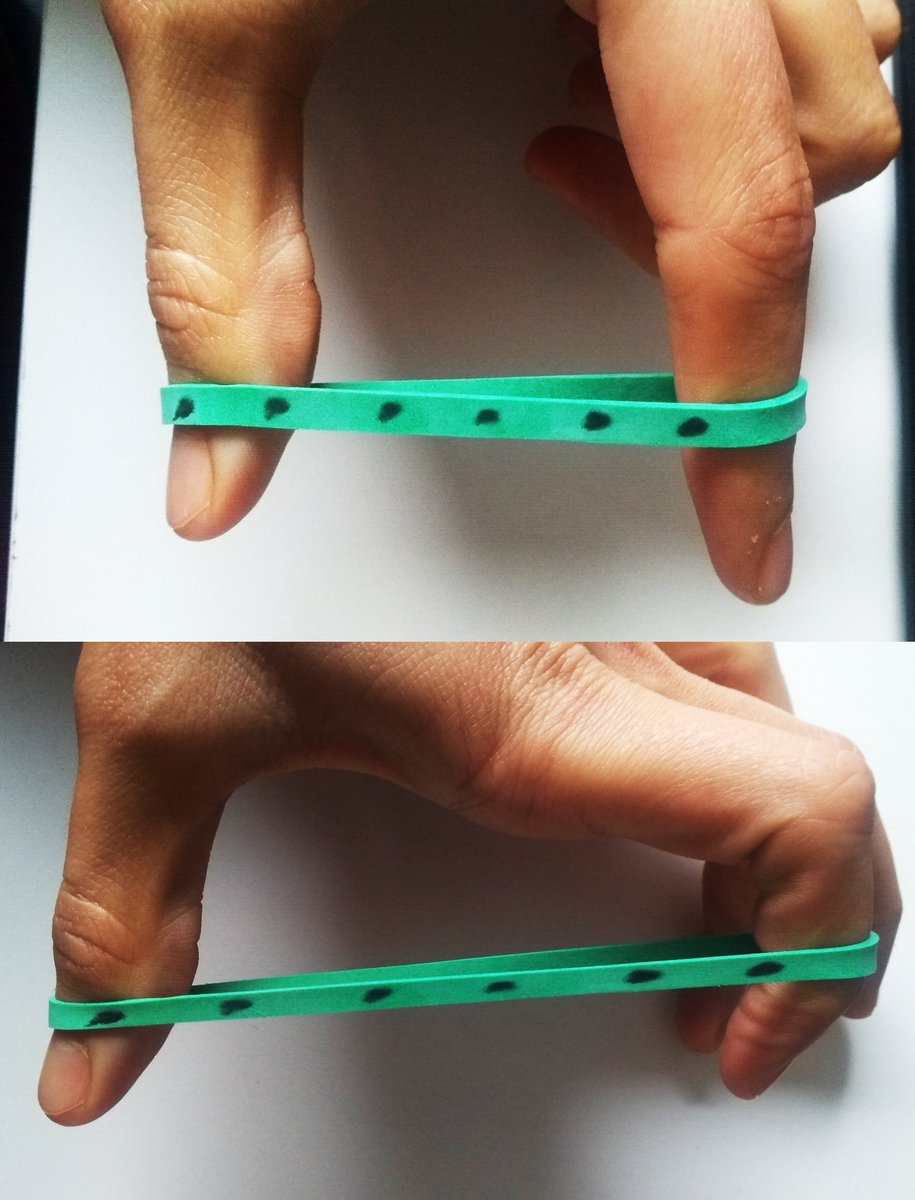
👀 Look at this dotted rubber band.
Let me give you an example.
👀 Look at this dotted rubber band.

(9/20) Now look at these other 2 images. I am applying a different force in each one.
If you compare them to the image of the previous tweet, can you tell in which one I am applying more force? 🧐
....
👏Easy, right? The more force I apply, the more the dots spread!
If you compare them to the image of the previous tweet, can you tell in which one I am applying more force? 🧐
....
👏Easy, right? The more force I apply, the more the dots spread!
(10/20) This is exactly what we do to measure cell forces!!
With the methodology #TractionForceMicroscopy, we place cells on an elastic gel (like the rubber before) with fluorescence beads inside. We take images before and after the cells apply forces and then we compare them!
With the methodology #TractionForceMicroscopy, we place cells on an elastic gel (like the rubber before) with fluorescence beads inside. We take images before and after the cells apply forces and then we compare them!

(11/20) In reality, these images have THOUSANDS of beads and also we need some math and theory of mechanics to calculate the forces from those images.
💻We need the help of computers and very complex algorithms to do these calculations!
AND there is one more problem...
💻We need the help of computers and very complex algorithms to do these calculations!
AND there is one more problem...
(12/20) In the last decades researchers are trying to make their #invitro cultures more similar to the human body. We have three-dimensional bodies, right? So, we also need to have #3D in vitro models!
So now we need to put our cells inside a gel, not on top!
So now we need to put our cells inside a gel, not on top!

(13/20) Did I mention that the algorithms needed are complex? Well, in 3D they become even more complex! 🤯
When I started my #PhD, there were few algorithms for 3D. And none were made available for any non-technical researcher to easily use them...
the time had come...
When I started my #PhD, there were few algorithms for 3D. And none were made available for any non-technical researcher to easily use them...
the time had come...
(14/20) In my #PhD, I developed an #opensource toolbox called #TFMLAB!
👨🔬👩🔬The main goal was that any researcher, regardless of their technical (💻math, programming, etc) experience, can easily use it!
It is user-friendly, intuitive and you don't need to know any programming!
👨🔬👩🔬The main goal was that any researcher, regardless of their technical (💻math, programming, etc) experience, can easily use it!
It is user-friendly, intuitive and you don't need to know any programming!
(15/20) #TFMLAB basically includes has all the complex algorithms that are needed to visualize the forces that cells exert in a given experiment! And again, it's #opensource ! You can find +info in this @ESBiomech tutorial:
Let's now look inside #TFMLAB🔍
Let's now look inside #TFMLAB🔍
(16/20) In my thesis we also developed a novel algorithm for #TractionForceMicroscopy valid for 3D and we tested it!
🏃Imagine that you want to run a mountain marathon but you don't train! You'd feel exhausted very soon!
That's why we trained our algorithm in a mountain!🏔️
🏃Imagine that you want to run a mountain marathon but you don't train! You'd feel exhausted very soon!
That's why we trained our algorithm in a mountain!🏔️
(17/20) We simulated conditions that the algorithm would need to face in a real experiment.
🔬For example, we tested it on images that look very much like the ones obtained with a microscope:
🔬For example, we tested it on images that look very much like the ones obtained with a microscope:

(18/20) After simulating thousands of different conditions, we saw that our algorithm is more accurate than others that are typically used in the field!
Interestingly, we obtained this improvement simply by making our algorithm consistent with Newton laws!
Interestingly, we obtained this improvement simply by making our algorithm consistent with Newton laws!

(19/20) These developments allowed us to see for the first time the forces that cells need to exert to form a new blood vessel!
We now have a tool ready to investigate, for example, vascular diseases, such as the one that my colleague @apek_shapeti is studying: #CCM
We now have a tool ready to investigate, for example, vascular diseases, such as the one that my colleague @apek_shapeti is studying: #CCM

(20/20) ✅In my #PhD I developed accurate tools to extract useful #force information from cells while making them accessible to the community🫱🫲.
The collaborative work of @MAtrix_KULeuven that joined forces with #MechanobiologyLab of @unisevilla made my work way easier! 💪💚
The collaborative work of @MAtrix_KULeuven that joined forces with #MechanobiologyLab of @unisevilla made my work way easier! 💪💚
@threadreaderapp unroll
• • •
Missing some Tweet in this thread? You can try to
force a refresh