We all think we're one of a kind.
But sometimes, we come across someone who looks just like us!
A @CellReports study tested the DNA of "fake twins".
Guess what:
They also share 🧬DNA variants related to facial features & behavior 🤯
Surprised or not really? Let’s dig in🧵👇
But sometimes, we come across someone who looks just like us!
A @CellReports study tested the DNA of "fake twins".
Guess what:
They also share 🧬DNA variants related to facial features & behavior 🤯
Surprised or not really? Let’s dig in🧵👇

First, let’s see why this study might NOT surprise you.
Monozygotic twins share almost identical facial traits & the same DNA sequence. Therefore, looking-alike strangers could follow a similar pattern.
Still, looking-alike strangers are not twins! So we can’t know for sure if:
Monozygotic twins share almost identical facial traits & the same DNA sequence. Therefore, looking-alike strangers could follow a similar pattern.
Still, looking-alike strangers are not twins! So we can’t know for sure if:
a. they share more of their genome than random people
b. if yes to (a), how much they share & what would be the functional role of the genes on which such SNVs are
c. how about #multiomics similarity, such as DNA methylation or microbiome (different in monozygotic twins)?
b. if yes to (a), how much they share & what would be the functional role of the genes on which such SNVs are
c. how about #multiomics similarity, such as DNA methylation or microbiome (different in monozygotic twins)?
Now, let’s see what this paper did.
1️⃣ Identify Human doubles (doppelgängers): pairs of people with very similar facial features, such that one can be confused with the other.
The pairs were selected from the photographic database of the artist Francois Brunelle.
1️⃣ Identify Human doubles (doppelgängers): pairs of people with very similar facial features, such that one can be confused with the other.
The pairs were selected from the photographic database of the artist Francois Brunelle.
This artist has been collecting artwork on Doppelgängers from around the world since 1999.
For this study, the authors started off with assessing 32 pairs.
francoisbrunelle.com/webn/e-project…
For this study, the authors started off with assessing 32 pairs.
francoisbrunelle.com/webn/e-project…
2️⃣ Devise a measure of “facial likeness” by combining the results of three #Deeplearning algorithms, each of which provides a score from 0 to 1 as to how similar two faces are: 1 denotes the same face, 0 corresponds to two completely different faces👇
a. the deep CNN Custom-Net algorithm, designed for surveillance (hertasecurity.com)
b. MatConvNet algorithm, designed for facial classification (Vedaldi and Lenc,2015)
c. Microsoft Oxford Project face API, designed for generalized facial analysis (azure.microsoft.com/es-es/services…)
b. MatConvNet algorithm, designed for facial classification (Vedaldi and Lenc,2015)
c. Microsoft Oxford Project face API, designed for generalized facial analysis (azure.microsoft.com/es-es/services…)
It’s interesting to understand how such algorithms work. For example, these are the 27 face landmarks computed by the Microsoft algorithm, based on which image similarity is inferred. 

Unsurprisingly, 25 out of the 32 look-alike pairs were found to be correlated by at least 2 of the 3 softwares. 

The plot below shows the 16 of 32 pairs (50%) which scored consistently high on all algorithms (LALs). For comparison, MZ-twins are monozygotic twin faces and Non-LALs are faces of random people.
Clearly, the selected look-alikes are more similar than random, less than MZ twins.
Clearly, the selected look-alikes are more similar than random, less than MZ twins.

3️⃣ Genetic analyses performed on the 16 pairs:
- genome: SNP microarrays
- epigenome: methylation microarrays
- microbiome
Based on various clusterings of the SNP profiles, 9 out of the 16 pairs were considered to be “ultra look-alike”.
The rest of the paper focuses on these.
- genome: SNP microarrays
- epigenome: methylation microarrays
- microbiome
Based on various clusterings of the SNP profiles, 9 out of the 16 pairs were considered to be “ultra look-alike”.
The rest of the paper focuses on these.
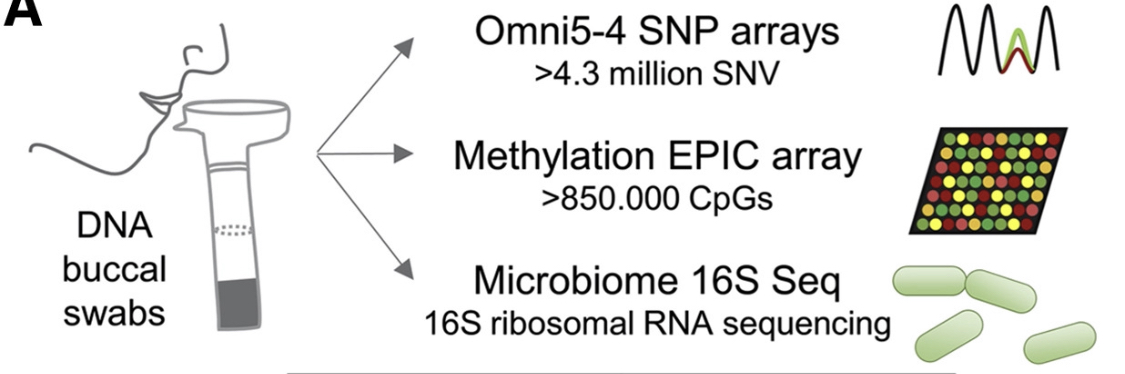
19,277 SNPs (located on 3,730 genes) were shared among the pairs.
Among those, 1,794 genes were “face-related” (as gathered from databases or prior GWAS results).
171 of the shared SNPs caused amino acid changes, affecting 158 genes, most of which related to facial determinants
Among those, 1,794 genes were “face-related” (as gathered from databases or prior GWAS results).
171 of the shared SNPs caused amino acid changes, affecting 158 genes, most of which related to facial determinants
4️⃣ Different epigenome and microbiome: even though they do share SNPs, the analyses in this paper concluded that Doppelgängers do not also share DNA methylation patterns or microbiome (at least not to a similar extent as SNPs).
5️⃣ Behavior: Behavioral features, extracted from biometric & lifestyle questionnaires, were used to calculate a likeness score of the 32 look-alike pairs.
Doppelgängers had higher likeness scores than the rest & higher similarity on particular behavioral features.
Doppelgängers had higher likeness scores than the rest & higher similarity on particular behavioral features.
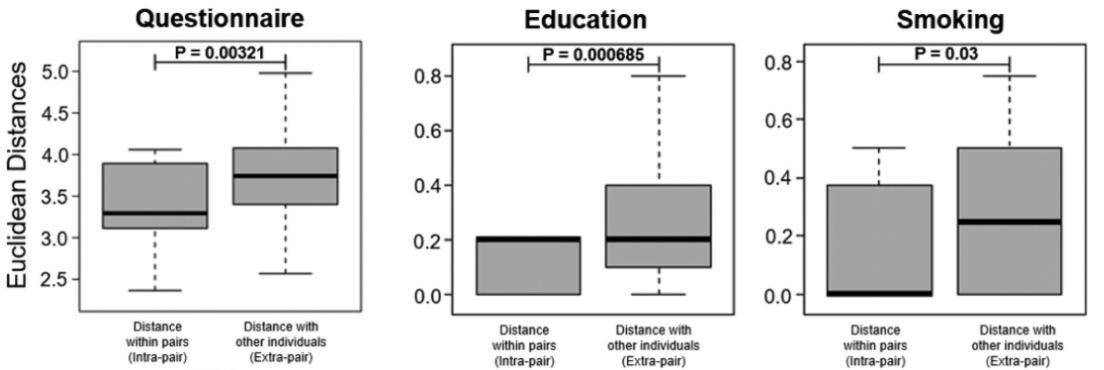
To me, it remains unclear though how robust these results are to changes in e.g. the metric used, the features on the basis of which the metric is computed, or increasing sample size (16 pairs is still low).
Robustly proving this behavioral link needs further analyses.
Robustly proving this behavioral link needs further analyses.
Let’s recap!
This paper looked at #multiomics of look-alike pairs, i.e. strangers with very similar facial features.
It found that they share more SNPs than expected (e.g. vs. random people) & that most SNPs relate to facial features.
No epigenome or microbiome sharing.
This paper looked at #multiomics of look-alike pairs, i.e. strangers with very similar facial features.
It found that they share more SNPs than expected (e.g. vs. random people) & that most SNPs relate to facial features.
No epigenome or microbiome sharing.
Some limitations of the study:
- small sample size
- more sensitivity analyses needed to increase robustness of results, s.a. varying more the metrics/cutoffs used
- potential selection biases by repeatedly shrinking down the already small sample size to highest signal
- small sample size
- more sensitivity analyses needed to increase robustness of results, s.a. varying more the metrics/cutoffs used
- potential selection biases by repeatedly shrinking down the already small sample size to highest signal
The dataset brought forward by this paper is unique, interesting & valuable.
The findings here are in line with the intuition that phenotypic similarities correlate with genomic ones and potentially also with behavior.
Data & custom scripts are openly available & downloadable.
The findings here are in line with the intuition that phenotypic similarities correlate with genomic ones and potentially also with behavior.
Data & custom scripts are openly available & downloadable.
Finally, here is the link to the @CellReports paper.
Congrats to all authors for this paper and for addressing this interesting question! @ManelEsteller @Alfons_Valencia
cell.com/cell-reports/f…
Congrats to all authors for this paper and for addressing this interesting question! @ManelEsteller @Alfons_Valencia
cell.com/cell-reports/f…
• • •
Missing some Tweet in this thread? You can try to
force a refresh